6TR6
 
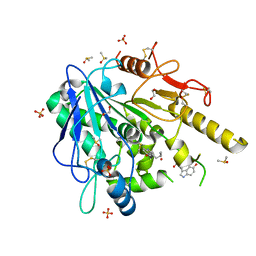 | | N-acetylserotonin-Notum complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of melatonin as an inhibitor of the Wnt deacylase Notum.
J. Pineal Res., 68, 2020
|
|
7MYP
 
 | | Crystal Structure of HIV-1 PRS17 with GRL-44-10A | | Descriptor: | (3R,3aS,4R,6aR)-4-methoxyhexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel HIV PR inhibitors with C4-substituted bis-THF and bis-fluoro-benzyl target the two active site mutations of highly drug resistant mutant PR S17 .
Biochem.Biophys.Res.Commun., 566, 2021
|
|
6YPW
 
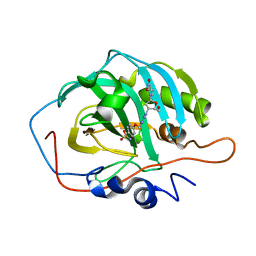 | | Crystal structure for the complex of human carbonic anhydrase II and 4-((1-(2-(hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-1H-1,2,3-triazol-4-yl)methoxy)benzenesulfonamide | | Descriptor: | 4-[[1-[(2~{S},3~{S},5~{R})-2-(hydroxymethyl)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]oxolan-3-yl]-1,2,3-triazol-4-yl]methoxy]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2020-04-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mechanisms of the Antiproliferative and Antitumor Activity of Novel Telomerase-Carbonic Anhydrase Dual-Hybrid Inhibitors.
J.Med.Chem., 64, 2021
|
|
8R8E
 
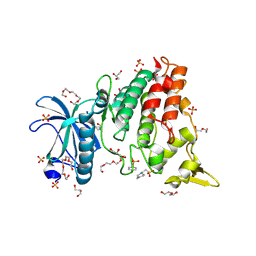 | | DYRK1a in Complex with 2-Cyclopentyl-7-iodo-1H-indole-3-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-7-iodanyl-1~{H}-indole-3-carbonitrile, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stahlecker, J, Dammann, M, Stehle, T, Boeckler, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2024-09-25 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Halogen Bonding on Water─A Drop in the Ocean?
J Chem Theory Comput, 20, 2024
|
|
4X14
 
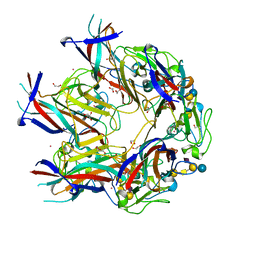 | | JC Mad-1 polyomavirus VP1 in complex with GM1 oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1, ... | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Greater Affinity of JC Polyomavirus Capsid for alpha 2,6-Linked Lactoseries Tetrasaccharide c than for Other Sialylated Glycans Is a Major Determinant of Infectivity.
J.Virol., 89, 2015
|
|
7N0U
 
 | | Complex of recombinant Bet v 1 with Fab fragment of REGN5713 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, REGN5713 Fab fragment heavy chain, REGN5713 Fab fragment light chain, ... | | Authors: | Franklin, M.C, Romero Hernandez, A. | | Deposit date: | 2021-05-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Targeting immunodominant Bet v 1 epitopes with monoclonal antibodies prevents the birch allergic response.
J.Allergy Clin.Immunol., 149, 2022
|
|
6AMP
 
 | | Crystal structure of H172A PHM (CuH absent, CuM present) | | Descriptor: | COPPER (II) ION, Peptidyl-glycine alpha-amidating monooxygenase | | Authors: | Maheshwari, S, Rudzka, K, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2017-08-10 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Effects of copper occupancy on the conformational landscape of peptidylglycine alpha-hydroxylating monooxygenase.
Commun Biol, 1, 2018
|
|
7RGN
 
 | | Crystal structure of putative fructose-1,6-bisphosphate aldolase from Candida auris | | Descriptor: | Fructose-bisphosphate aldolase, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative fructose-1,6-bisphosphate aldolase from Candida auris
To Be Published
|
|
9L78
 
 | |
8RJA
 
 | | Crystal structure of the F420-reducing formylmethanofuran dehydrogenase complex from the ethanotroph Candidatus Ethanoperedens thermophilum | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Lemaire, O.N, Wagner, T. | | Deposit date: | 2023-12-20 | | Release date: | 2024-10-02 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Ethane-oxidising archaea couple CO 2 generation to F 420 reduction.
Nat Commun, 15, 2024
|
|
9L6K
 
 | |
5N0D
 
 | | Crystal structure of human carbonic anhydrase II in complex with (R)-4-(6,7-dihydroxy-1-phenyl-3,4-tetrahydroisoquinoline-1H-2-carbonyl)benzenesulfonamide. | | Descriptor: | (R)-4-(6,7-dihydroxy-1-phenyl-3,4-tetrahydroisoquinoline-1H-2-carbonyl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing Molecular Interactions between Human Carbonic Anhydrases (hCAs) and a Novel Class of Benzenesulfonamides.
J. Med. Chem., 60, 2017
|
|
9L7M
 
 | | Nucleotide-free kinesin-1 motor domain bound to the microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, MAGNESIUM ION, ... | | Authors: | Makino, T, Komori, Y, Yanagisawa, H, Tomishige, M, Kikkawa, M. | | Deposit date: | 2024-12-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Tension-induced suppression of allosteric conformational changes coordinates kinesin-1 stepping.
J.Cell Biol., 224, 2025
|
|
8AOF
 
 | | Specific covalent inhibitor(16) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.615 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8AOA
 
 | | Covalent and non-covalent inhibitor of ERK2 (two sites) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
7B3H
 
 | | Notum complex with ARUK3003909 | | Descriptor: | 1,2-ETHANEDIOL, 6-((3-(trifluoromethoxy)phenyl)thio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
6QGH
 
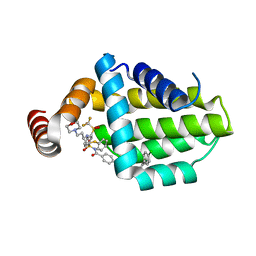 | | Structure of human Bcl-2 in complex with ABT-263 | | Descriptor: | 4-(4-{[2-(4-chlorophenyl)-5,5-dimethylcyclohex-1-en-1-yl]methyl}piperazin-1-yl)-N-[(4-{[(2R)-4-(morpholin-4-yl)-1-(phenylsulfanyl)butan-2-yl]amino}-3-[(trifluoromethyl)sulfonyl]phenyl)sulfonyl]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QGJ
 
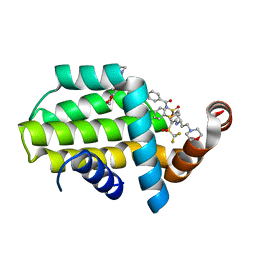 | | Structure of human Bcl-2 in complex with fragment/ABT-263 hybrid | | Descriptor: | 4-[4-[(1~{R})-1-(6-methoxy-1,3-benzodioxol-5-yl)-2-pyrrolidin-1-yl-ethyl]phenyl]-~{N}-[4-[[(2~{R})-4-morpholin-4-yl-1-phenylsulfanyl-butan-2-yl]amino]-3-(trifluoromethylsulfonyl)phenyl]sulfonyl-benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
7B3P
 
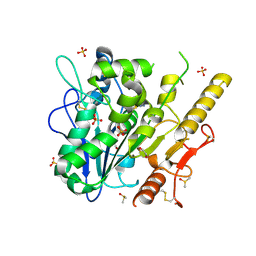 | | Notum complex with ARUK3003775 | | Descriptor: | 1,2-ETHANEDIOL, 6-((4-chlorophenyl)thio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
7B2N
 
 | | Crystal structure of Chlamydomonas reinhardtii chloroplastic Fructose bisphosphate aldolase | | Descriptor: | CHLORIDE ION, Fructose-bisphosphate aldolase 1, chloroplastic, ... | | Authors: | Le Moigne, T, Lemaire, S.D, Henri, J. | | Deposit date: | 2020-11-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of chloroplast fructose-1,6-bisphosphate aldolase from the green alga Chlamydomonas reinhardtii.
J.Struct.Biol., 214, 2022
|
|
7B3G
 
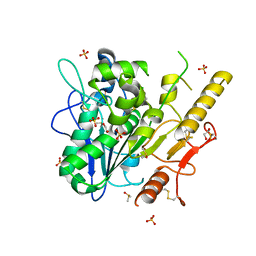 | | Notum complex with ARUK3003902 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-((2-chlorophenyl)thio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, ... | | Authors: | Zhao, Y, Jone, E.Y. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
8RIK
 
 | | Microtubule-associated kinesin-1 tail complex bound to ADP, single-headed state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Atherton, J, Chegkazi, M.S, Steiner, R.A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Microtubule association induces a Mg-free apo-like ADP pre-release conformation in kinesin-1 that is unaffected by its autoinhibitory tail.
Nat Commun, 16, 2025
|
|
7B8C
 
 | | Notum-Fragment 147 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-~{N}-(2-methylpropyl)imidazo[1,2-a]pyridine-3-carboxamide, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B8M
 
 | | Notum-Fragment 193 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methoxy-2-(1~{H}-pyrazol-3-yl)phenol, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-13 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
7B84
 
 | | Notum-Fragment065 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jonees, E.Y. | | Deposit date: | 2020-12-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
