2RRQ
 
 | | DNA oligomer containing propylene cross-linked cyclic 2'-deoxyuridylate dimer | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*CP*AP*(JDT)P*TP*AP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*GP*TP*AP*AP*TP*GP*AP*AP*GP*G)-3') | | Authors: | Furuita, K, Murata, S, Jee, J.G, Ichikawa, S, Matsuda, A, Kojima, C. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Feature of Bent DNA Recognized by HMGB1
J.Am.Chem.Soc., 133, 2011
|
|
5ZGF
 
 | | Crystal structure of NDM-1 Q123G mutant | | Descriptor: | HYDROXIDE ION, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Hao, Q. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Active-Site Conformational Fluctuations Promote the Enzymatic Activity of NDM-1.
Antimicrob. Agents Chemother., 62, 2018
|
|
5ZGI
 
 | |
5ZKE
 
 | | Crystal Structure of N-terminal Domain of Plasmodium vivax p43 in space group P212121 | | Descriptor: | Aminoacyl tRNA Synthetase Complex-Interacting Multifunctional Protein p43 | | Authors: | Gupta, S, Sharma, M, Harlos, K, Manickam, Y, Sharma, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.492 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5ZLI
 
 | |
2RUT
 
 | | Solution structures of the DNA-binding domain (ZF2) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT.
J Struct Funct Genomics, 16, 2015
|
|
2RKV
 
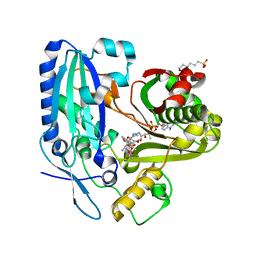 | | Crystal Structure of F. graminearum TRI101 complexed with Coenzyme A and T-2 mycotoxin | | Descriptor: | 12,13-Epoxytrichothec-9-ene-3,4,8,15-tetrol-4,15-diacetate-8-isovalerate, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, COENZYME A, ... | | Authors: | Garvey, G.S, Rayment, I. | | Deposit date: | 2007-10-17 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of the TRI101 Trichothecene 3-O-Acetyltransferase from Fusarium sporotrichioides and Fusarium graminearum: KINETIC INSIGHTS TO COMBATING FUSARIUM HEAD BLIGHT
J.Biol.Chem., 283, 2008
|
|
2RUY
 
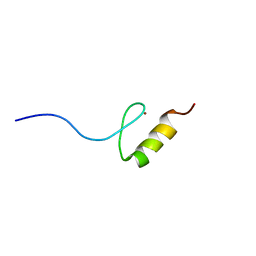 | | Solution structures of the DNA-binding domain (ZF10) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
7B38
 
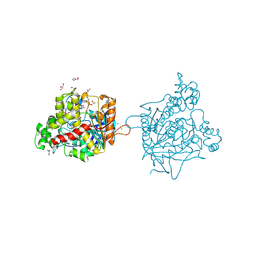 | | Torpedo californica acetylcholinesterase complexed with Mg+2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
7B8E
 
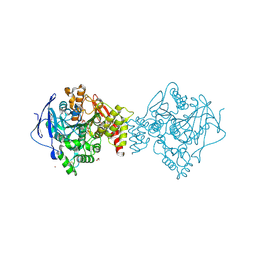 | | Torpedo californica acetylcholinesterase complexed with Ca+2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L. | | Deposit date: | 2020-12-12 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
2ROU
 
 | | Stereospecific Conformations of N2-dG 1R-trans-anti-Benzo[c]phenanthrene DNA Adducts: 3'-Intercalation of the 1R Adduct and 5'-Minor Groove Orientation of the 1S Adduct in an Iterated (CG)3 Repeat | | Descriptor: | (1R)-1,2,3,4-TETRAHYDRO-BENZO[C]PHENANTHRENE-2,3,4-TRIOL, DNA (5'-D(*DAP*DTP*DCP*DGP*DCP*DGP*DCP*DGP*DGP*DCP*DAP*DTP*DG)-3'), DNA (5'-D(*DCP*DAP*DTP*DGP*DCP*DCP*DGP*DCP*DGP*DCP*DGP*DAP*DT)-3') | | Authors: | Wang, Y, Kroth, H, Yagi, H, Sayer, J.M, Kumar, S, Jerina, D.M, Stone, M.P. | | Deposit date: | 2008-04-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 3'-Intercalation of a N2-dG 1R-trans-anti-benzo[c]phenanthrene DNA adduct in an iterated (CG)3 repeat
Chem.Res.Toxicol., 21, 2008
|
|
5ZMA
 
 | | Structural basis for an allosteric Eya2 phosphatase inhibitor | | Descriptor: | 3-fluoro-N'-[(E)-{5-[(pyrimidin-2-yl)sulfanyl]furan-2-yl}methylidene]benzohydrazide, Eyes absent homolog 2 | | Authors: | Anantharajan, J, Jansson, A.E, Kang, C. | | Deposit date: | 2018-04-02 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.175 Å) | | Cite: | Structural and Functional Analyses of an Allosteric EYA2 Phosphatase Inhibitor That Has On-Target Effects in Human Lung Cancer Cells.
Mol.Cancer Ther., 18, 2019
|
|
2RPD
 
 | | Mhr1p-bound ssDNA | | Descriptor: | DNA (5'-D(*DTP*DAP*DCP*DG)-3') | | Authors: | Masuda, T, Ito, Y, Shibata, T, Mikawa, T. | | Deposit date: | 2008-05-15 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A non-canonical DNA structure enables homologous recombination in various genetic systems
J.Biol.Chem., 284, 2009
|
|
5ZMO
 
 | | Sulfur binding domain of ScoMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(P*CP*CP*GP*(GS)P*CP*CP*GP*G)-3'), PHOSPHATE ION, Uncharacterized protein McrA | | Authors: | Liu, G, Fu, W, Zhang, Z, He, Y, Yu, H, Zhao, Y, Deng, Z, Wu, G, He, X. | | Deposit date: | 2018-04-04 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for the recognition of sulfur in phosphorothioated DNA.
Nat Commun, 9, 2018
|
|
5ZN7
 
 | |
2UP1
 
 | | STRUCTURE OF UP1-TELOMERIC DNA COMPLEX | | Descriptor: | DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), PROTEIN (HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1) | | Authors: | Ding, J, Hayashi, M.K, Krainer, A.R, Xu, R.-M. | | Deposit date: | 1998-07-10 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA.
Genes Dev., 13, 1999
|
|
5ZNI
 
 | |
5ZQ8
 
 | | Crystal structure of spRlmCD with U747 stemloop RNA | | Descriptor: | NICKEL (II) ION, RNA (5'-R(*CP*CP*GP*UP*(MUM)P*GP*AP*AP*AP*AP*GP*G)-3'), S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Jiang, Y.Y, Yu, H.L. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Unveiling the structural features that determine the dual methyltransferase activities of Streptococcus pneumoniae RlmCD
PLoS Pathog., 14, 2018
|
|
2RUX
 
 | | Solution structures of the DNA-binding domain (ZF6) of immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RV4
 
 | | Solution structures of the DNA-binding domain (ZF5) of mouse immune-related zinc-finger protein ZFAT | | Descriptor: | ZINC ION, Zinc finger protein ZFAT | | Authors: | Tochio, N, Umehara, T, Kigawa, T, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the DNA-binding domains of immune-related zinc-finger protein ZFAT
J.Struct.Funct.Genom., 16, 2015
|
|
2RFK
 
 | | Substrate RNA Positioning in the Archaeal H/ACA Ribonucleoprotein Complex | | Descriptor: | Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, Small nucleolar rnp similar to gar1, ... | | Authors: | Liang, B, Xue, S, Terns, R.M, Terns, M.P, Li, H, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-09-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2RGA
 
 | | Crystal structure of H-RasQ61I-GppNHp | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Buhrman, G, Wink, G, Mattos, C. | | Deposit date: | 2007-10-03 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transformation Efficiency of RasQ61 Mutants Linked to Structural Features of the Switch Regions in the Presence of Raf.
Structure, 15, 2007
|
|
2RHD
 
 | | Crystal structure of Cryptosporidium parvum small GTPase RAB1A | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Small GTP binding protein rab1a | | Authors: | Dong, A, Xu, X, Lew, J, Lin, Y.H, Khuu, C, Sun, X, Qiu, W, Kozieradzki, I, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Sukumar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of Cryptosporidium parvum small GTPase RAB1A.
To be Published
|
|
5ZB6
 
 | |
5ZRD
 
 | | Tyrosinase from Burkholderia thailandensis (BtTYR) at low pH condition | | Descriptor: | CITRIC ACID, COPPER (II) ION, GLYCEROL, ... | | Authors: | Lee, S, Son, H.-F, Kim, K.-J. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Highly Efficient Production of Catechol Derivatives at Acidic pH by Tyrosinase from Burkholderia thailandensis
Acs Catalysis, 8, 2018
|
|
