3U99
 
 | |
1CIE
 
 | |
3OKR
 
 | |
3Q3Z
 
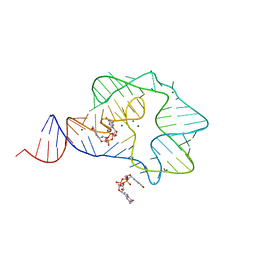 | | Structure of a c-di-GMP-II riboswitch from C. acetobutylicum bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, c-di-GMP-II riboswitch | | Authors: | Smith, K.D, Shanahan, C.A, Moore, E.L, Simon, A.C, Strobel, S.A. | | Deposit date: | 2010-12-22 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of differential ligand recognition by two classes of bis-(3'-5')-cyclic dimeric guanosine monophosphate-binding riboswitches.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1CIH
 
 | |
1PK8
 
 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-06-05 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
1CSX
 
 | |
1CSW
 
 | |
1CSV
 
 | |
1CSU
 
 | |
1CIF
 
 | |
2BSA
 
 | | Ferredoxin-Nadp Reductase (Mutation: Y 303 S) complexed with NADP | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maya, C.M, Hermoso, J.A, Perez-Dorado, I, Tejero, J, Julvez, M.M, Gomez-Moreno, C, Medina, M. | | Deposit date: | 2005-05-20 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
1EGG
 
 | | STRUCTURE OF A C-TYPE CARBOHYDRATE-RECOGNITION DOMAIN (CRD-4) FROM THE MACROPHAGE MANNOSE RECEPTOR | | Descriptor: | CALCIUM ION, MACROPHAGE MANNOSE RECEPTOR | | Authors: | Feinberg, H, Park-Snyder, S, Kolatkar, A.R, Heise, C.T, Taylor, M.E, Weis, W.I. | | Deposit date: | 2000-02-15 | | Release date: | 2000-08-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a C-type carbohydrate recognition domain from the macrophage mannose receptor.
J.Biol.Chem., 275, 2000
|
|
2HL7
 
 | | Crystal structure of the periplasmic domain of CcmH from Pseudomonas aeruginosa | | Descriptor: | Cytochrome C-type biogenesis protein CcmH, TETRAETHYLENE GLYCOL | | Authors: | Di Matteo, A, Travaglini-Allocatelli, C, Gianni, S, Brunori, M. | | Deposit date: | 2006-07-06 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A strategic protein in cytochrome c maturation: three-dimensional structure of CcmH and binding to apocytochrome c
J.Biol.Chem., 282, 2007
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
3ZQC
 
 | | Structure of the Trichomonas vaginalis Myb3 DNA-binding domain bound to a promoter sequence reveals a unique C-terminal beta-hairpin conformation | | Descriptor: | MRE-1, MYB3 | | Authors: | Wei, S.-Y, Lou, Y.-C, Tsai, J.-Y, Hsu, H.-M, Tai, J.-H, Hsiao, C.-D, Chen, C. | | Deposit date: | 2011-06-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Trichomonas Vaginalis Myb3 DNA-Binding Domain Bound to a Promoter Sequence Reveals a Unique C-Terminal Beta-Hairpin Conformation.
Nucleic Acids Res., 40, 2012
|
|
1QMD
 
 | | calcium bound closed form alpha-toxin from Clostridium perfringens | | Descriptor: | CALCIUM ION, PHOSPHOLIPASE C, ZINC ION | | Authors: | Naylor, C.E, Miller, J, Titball, R.W, Basak, A.K. | | Deposit date: | 1999-09-27 | | Release date: | 2000-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterisation of the Calcium-Binding C-Terminal Domain of Clostridium Perfringens Alpha-Toxin
J.Mol.Biol., 294, 1999
|
|
1QM6
 
 | | Closed form of Clostridium perfringens alpha-toxin strain NCTC8237 | | Descriptor: | PHOSPHOLIPASE C, ZINC ION | | Authors: | Naylor, C.E, Miller, J, Titball, R.W, Basak, A.K. | | Deposit date: | 1999-09-21 | | Release date: | 1999-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterisation of the Calcium-Binding C-Terminal Domain of Clostridium Perfringens Alpha-Toxin
J.Mol.Biol., 294, 1999
|
|
3OUQ
 
 | | Structure of N-terminal hexaheme fragment of GSU1996 | | Descriptor: | Cytochrome c family protein, HEME C | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a novel dodecaheme cytochrome c from Geobacter sulfurreducens reveals an extended 12nm protein with interacting hemes.
J.Struct.Biol., 174, 2011
|
|
2F55
 
 | | Two hepatitis c virus ns3 helicase domains complexed with the same strand of dna | | Descriptor: | 5'-D(P*(DU)P*(DU)P*(DU))-3', 5'-D(P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU)P*(DU))-3', SULFATE ION, ... | | Authors: | Lu, J.Z, Jordan, J.B, Sakon, J. | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and biological identification of residues on the surface of NS3 helicase required for optimal replication of the hepatitis C virus
J.Biol.Chem., 281, 2006
|
|
5TRY
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2206 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-3-methoxy-1-(naphthalen-1-ylmethylamino)-1-oxidanylidene-propan-2-yl]-4-oxidanylidene-2-(3-phenylpropanoylamino)-4-piperidin-1-yl-butanamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.000008 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
2F42
 
 | |
2UZP
 
 | | Crystal structure of the C2 domain of human protein kinase C gamma. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Pike, A.C.W, Amos, A, Johansson, C, Sobott, F, Savitsky, P, Berridge, G, Fedorov, O, Umeano, C, Gorrec, F, Bunkoczi, G, Debreczeni, J, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of C2 Domain of Protein Kinase C Gamma
To be Published
|
|
5TS0
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2208 | | Descriptor: | (2S)-N-{(2S)-3-methoxy-1-[(naphthalen-1-ylmethyl)amino]-1-oxopropan-2-yl}-4-oxo-2-[(3-phenylpropanoyl)amino]-4-(1H-pyrrol-1-yl)butanamide (non-preferred name), Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.84679747 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5TRS
 
 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide PKS2144 | | Descriptor: | N-tert-butoxy-N~2~-(5-methyl-1,2-oxazole-3-carbonyl)-L-asparaginyl-O-methyl-N-[(naphthalen-1-yl)methyl]-L-serinamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, P.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.083567 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
