3TNQ
 
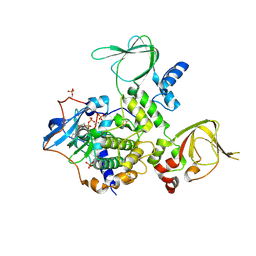 | | Structure and Allostery of the PKA RIIb Tetrameric Holoenzyme | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein kinase, ... | | Authors: | Zhang, P, Smith-Nguyen, E.V, Keshwani, M.M, Deal, M.S, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-01 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structure and allostery of the PKA RIIbeta tetrameric holoenzyme
Science, 335, 2012
|
|
3TPM
 
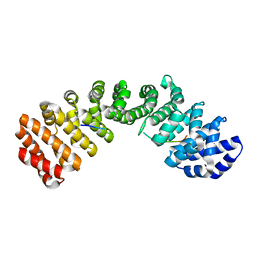 | |
8P8X
 
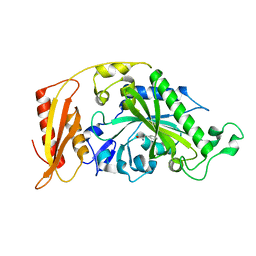 | |
8JH4
 
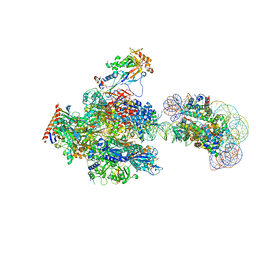 | | RNA polymerase II elongation complex containing 60 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH2
 
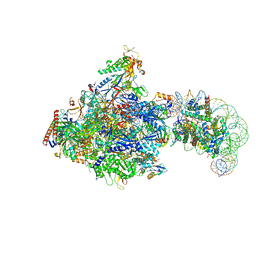 | | RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (218-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH3
 
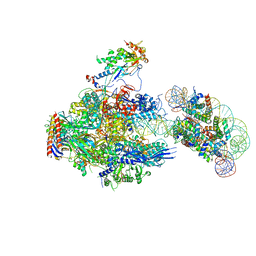 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
3TNP
 
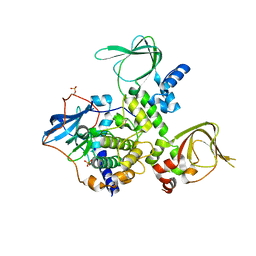 | | Structure and Allostery of the PKA RIIb Tetrameric Holoenzyme | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Zhang, P, Smith-Nguyen, E.V, Keshwani, M.M, Deal, M.S, Kornev, A.P, Taylor, S.S. | | Deposit date: | 2011-09-01 | | Release date: | 2012-02-01 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and allostery of the PKA RIIbeta tetrameric holoenzyme
Science, 335, 2012
|
|
3RSK
 
 | |
258L
 
 | | AN ADAPTABLE METAL-BINDING SITE ENGINEERED INTO T4 LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME, ZINC ION | | Authors: | Wray, J.W, Baase, W.A, Ostheimer, G.J, Matthews, B.W. | | Deposit date: | 1999-01-05 | | Release date: | 2000-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of a non-rigid region in T4 lysozyme to design an adaptable metal-binding site.
Protein Eng., 13, 2000
|
|
7DLY
 
 | | Crystal structure of Arabidopsis ACS7 mutant in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7 | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
7DLW
 
 | | Crystal structure of Arabidopsis ACS7 in complex with PPG | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, 1-aminocyclopropane-1-carboxylate synthase 7, SULFATE ION | | Authors: | Hao, B, Zhang, Y, Li, X, Rao, Z. | | Deposit date: | 2020-11-30 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Dual activities of ACC synthase: Novel clues regarding the molecular evolution of ACS genes.
Sci Adv, 7, 2021
|
|
210L
 
 | | PROTEIN STRUCTURE PLASTICITY EXEMPLIFIED BY INSERTION AND DELETION MUTANTS IN T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, T4 LYSOZYME | | Authors: | Vetter, I.R, Baase, W.A, Heinz, D.W, Xiong, J.-P, Snow, S, Matthews, B.W. | | Deposit date: | 1996-09-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Protein structural plasticity exemplified by insertion and deletion mutants in T4 lysozyme.
Protein Sci., 5, 1996
|
|
3RYH
 
 | | GMPCPP-Tubulin: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Nawrotek, A, Knossow, M, Gigant, B. | | Deposit date: | 2011-05-11 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Determinants That Govern Microtubule Assembly from the Atomic Structure of GTP-Tubulin.
J.Mol.Biol., 412, 2011
|
|
8IQY
 
 | |
8IQW
 
 | |
3SLN
 
 | | Structural characterization of a GII.4 2004 norovirus variant (TCH05) bound to H pentasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shanker, S, Choi, J.-M, Sankaran, B, Atmar, R.L, Estes, M.K, Prasad, B.V.V. | | Deposit date: | 2011-06-24 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structural Analysis of Histo-Blood Group Antigen Binding Specificity in a Norovirus GII.4 Epidemic Variant: Implications for Epochal Evolution.
J.Virol., 85, 2011
|
|
3RYF
 
 | | GTP-Tubulin: RB3 Stathmin-like domain complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Nawrotek, A, Knossow, M, Gigant, B. | | Deposit date: | 2011-05-11 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Determinants That Govern Microtubule Assembly from the Atomic Structure of GTP-Tubulin.
J.Mol.Biol., 412, 2011
|
|
3U0T
 
 | | Fab-antibody complex | | Descriptor: | Amyloid beta A4 protein, ponezumab HC Fab, ponezumab LC Fab | | Authors: | LaPorte, S.L, Pons, J.P. | | Deposit date: | 2011-09-29 | | Release date: | 2012-01-11 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of C-terminal beta-Amyloid Peptide Binding by the Antibody Ponezumab for the Treatment of Alzheimer's Disease
J.Mol.Biol., 421, 2012
|
|
3KNX
 
 | | HCV NS3 protease domain with P1-P3 macrocyclic ketoamide inhibitor | | Descriptor: | (2R)-2-{(3S,13S,16aS,17aR,17bS)-13-[({(1S)-1-[(4,4-dimethyl-2,6-dioxopiperidin-1-yl)methyl]-2,2-dimethylpropyl}carbamoyl)amino]-17,17-dimethyl-1,14-dioxooctadecahydro-2H-cyclopropa[3,4]pyrrolo[1,2-a][1,4]diazacyclohexadecin-3-yl}-2-hydroxy-N-prop-2-en-1-ylethanamide, BETA-MERCAPTOETHANOL, HCV NS3 Protease, ... | | Authors: | Venkatraman, S, Velazquez, F, Wu, W, Blackman, M, Chen, K.X, Bogen, S, Nair, L, Tong, X, Chase, R, Hart, A, Agrawal, S, Pichardo, J, Prongay, A, Cheng, K.-C, Girijavallabhan, V, Piwinski, J, Shih, N.-Y, Njoroge, F.G. | | Deposit date: | 2009-11-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and structure-activity relationship of P1-P3 ketoamide derived macrocyclic inhibitors of hepatitis C virus NS3 protease.
J.Med.Chem., 52, 2009
|
|
3TPQ
 
 | |
2C77
 
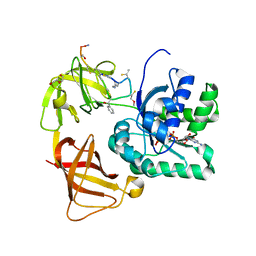 | | EF-Tu complexed with a GTP analog and the antibiotic GE2270 A | | Descriptor: | DI(HYDROXYETHYL)ETHER, ELONGATION FACTOR TU-B, MAGNESIUM ION, ... | | Authors: | Parmeggiani, A, Krab, I.M, Okamura, S, Nielsen, R.C, Nyborg, J, Nissen, P. | | Deposit date: | 2005-11-18 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu.
Biochemistry, 45, 2006
|
|
3QM4
 
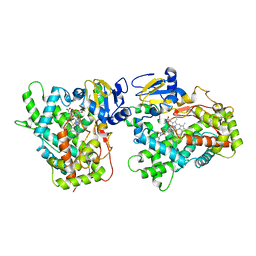 | | Human Cytochrome P450 (CYP) 2D6 - Prinomastat Complex | | Descriptor: | Cytochrome P450 2D6, NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Human Cytochrome P450 2D6 with Prinomastat Bound.
J.Biol.Chem., 287, 2012
|
|
3QNG
 
 | |
3QNW
 
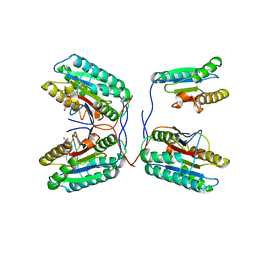 | | Caspase-6 in complex with Z-VAD-FMK inhibitor | | Descriptor: | Caspase-6, Z-VAD-FMK | | Authors: | Mueller, I, Lamers, M, Ritchie, A, Park, H, Dominguez, C, Munoz, I, Maillard, M, Kiselyov, A. | | Deposit date: | 2011-02-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of human caspase-6 in complex with Z-VAD-FMK: New peptide binding mode observed for the non-canonical caspase conformation.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1XYF
 
 | | ENDO-1,4-BETA-XYLANASE FROM STREPTOMYCES OLIVACEOVIRIDIS | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Fujimoto, Z, Mizuno, H, Kuno, A, Kusakabe, I. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Streptomyces olivaceoviridis E-86 beta-xylanase containing xylan-binding domain.
J.Mol.Biol., 300, 2000
|
|
