7TDJ
 
 | | Rabbit RyR1 with AMP-PCP and high Ca2+ embedded in nanodisc in closed-inactivated conformation class 1(Dataset-A) | | Descriptor: | CALCIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Ryanodine receptor 1,Ryanodine receptor 1,RyR1, ... | | Authors: | Nayak, A.R, Samso, M. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ca 2+ -inactivation of the mammalian ryanodine receptor type 1 in a lipidic environment revealed by cryo-EM.
Elife, 11, 2022
|
|
7TDG
 
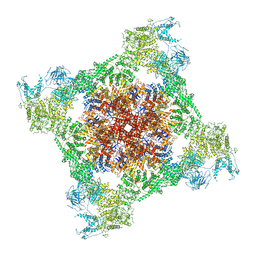 | |
4OAV
 
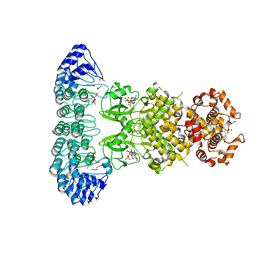 | | Complete human RNase L in complex with 2-5A (5'-ppp heptamer), AMPPCP and RNA substrate. | | Descriptor: | (2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-4-hydroxy-2-({[(S)-hydroxy{[(2R,3S,4S)-4-hydroxy-2-(hydroxymethyl)tetrahydrofuran-3-yl]oxy}phosphoryl]oxy}methyl)tetrahydrofuran-3-yl dihydrogen phosphate, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Han, Y, Donovan, J, Rath, S, Whitney, G, Chitrakar, A, Korennykh, A. | | Deposit date: | 2014-01-06 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response.
Science, 343, 2014
|
|
4M0K
 
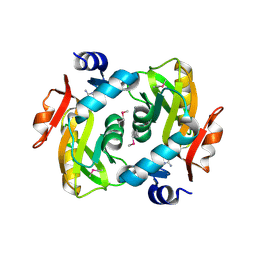 | | Crystal structure of adenine phosphoribosyltransferase from Rhodothermus marinus DSM 4252, NYSGRC Target 029775. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, CALCIUM ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of adenine phosphoribosyltransferase from Rhodothermus marinus DSM 4252, NYSGRC Target 029775.
To be Published
|
|
4FLC
 
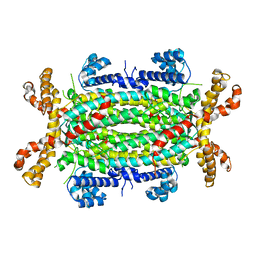 | | Structural and Biochemical Characterization of Human Adenylosuccinate Lyase (ADSL) and the R303C ADSL Deficiency Associated Mutation | | Descriptor: | Adenylosuccinate lyase | | Authors: | Deaton, M.K, Ray, S.P, Capodagli, G.C, Calkins, L.A.F, Sawle, L, Ghosh, K, Patterson, D, Pegan, S.D. | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Biochemical Characterization of Human Adenylosuccinate Lyase (ADSL) and the R303C ADSL Deficiency-Associated Mutation.
Biochemistry, 51, 2012
|
|
1NOJ
 
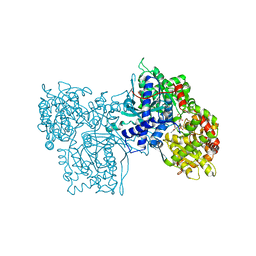 | |
1NOI
 
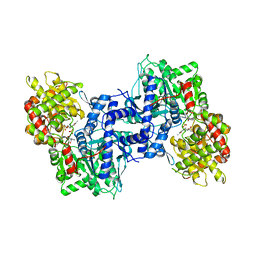 | |
1NOK
 
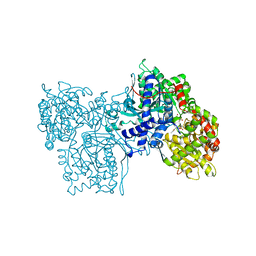 | |
8WR8
 
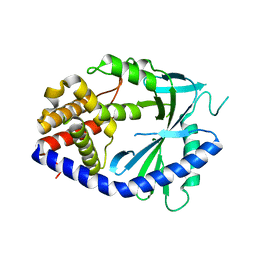 | |
1E24
 
 | | LYSYL-TRNA SYNTHETASE (LYSU) HEXAGONAL FORM complexed with lysine and ATP and MN2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, LYSINE, ... | | Authors: | Desogus, G, Todone, F, Brick, P, Onesti, S. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Active Site of Lysyl-tRNA Synthetase: Structural Studies of the Adenylation Reaction
Biochemistry, 39, 2000
|
|
1E1T
 
 | | LYSYL-TRNA SYNTHETASE (LYSU) HEXAGONAL FORM COMPLEXED WITH THE LYSYL_ADENYLATE INTERMEDIATE | | Descriptor: | ADENOSINE-5'-[LYSYL-PHOSPHATE], GLYCEROL, LYSYL-TRNA SYNTHETASE, ... | | Authors: | Desogus, G, Todone, F, Brick, P, Onesti, S. | | Deposit date: | 2000-05-10 | | Release date: | 2000-07-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active Site of Lysyl-tRNA Synthetase: Structural Studies of the Adenylation Reaction
Biochemistry, 39, 2000
|
|
1E1O
 
 | | lysyl-tRNA Synthetase (LYSU) hexagonal form, complexed with lysine | | Descriptor: | GLYCEROL, LYSINE, LYSYL-TRNA SYNTHETASE, ... | | Authors: | Desogus, G, Todone, F, Brick, P, Onesti, S. | | Deposit date: | 2000-05-09 | | Release date: | 2000-07-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Active Site of Lysyl-tRNA Synthetase: Structural Studies of the Adenylation Reaction
Biochemistry, 39, 2000
|
|
1ULT
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | CITRIC ACID, long chain fatty acid-CoA ligase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-16 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of the substrate specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer
J.Biol.Chem., 279, 2004
|
|
1V25
 
 | | Crystal structure of tt0168 from Thermus thermophilus HB8 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, long-chain-fatty-acid-CoA synthetase | | Authors: | Hisanaga, Y, Ago, H, Nakatsu, T, Hamada, K, Ida, K, Kanda, H, Yamamoto, M, Hori, T, Arii, Y, Sugahara, M, Kuramitsu, S, Yokoyama, S, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-07 | | Release date: | 2004-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Substrate-specific Two-step Catalysis of Long Chain Fatty Acyl-CoA Synthetase Dimer
J.Biol.Chem., 279, 2004
|
|
2D1S
 
 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1T
 
 | | Crystal structure of the thermostable Japanese Firefly Luciferase red-color emission S286N mutant complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2YQ9
 
 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation V321A, crystallized with 2'-AMP | | Descriptor: | 2', 3'-CYCLIC-NUCLEOTIDE 3'-PHOSPHODIESTERASE, ADENOSINE-2'-MONOPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Lehtimaki, M, Han, H, Kursula, P. | | Deposit date: | 2012-11-07 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Analysis of the Reaction Cycle of 2',3'-Cyclic Nucleotide 3'-Phosphodiesterase, a Unique Member of the 2H Phosphoesterase Family
J.Mol.Biol., 425, 2013
|
|
4UJ2
 
 | | Protein Kinase A in complex with an Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(3S,4R)-4-(3-iodanylphenyl)carbonylpyrrolidin-3-yl]-3H-quinazolin-4-one, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of Pka and a Pkb Mimic.
Chemistry, 22, 2016
|
|
3SHR
 
 | | Crystal Structure of cGMP-dependent Protein Kinase Reveals Novel Site of Interchain Communication | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, SULFATE ION, cGMP-dependent protein kinase 1 | | Authors: | Osborne, B.W, Wu, J, Taylor, S.S, Dostmann, W.R. | | Deposit date: | 2011-06-16 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of cGMP-Dependent Protein Kinase Reveals Novel Site of Interchain Communication.
Structure, 19, 2011
|
|
4UJ9
 
 | | Protein Kinase A in complex with an Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(3S,4R)-4-[4-(trifluoromethyl)phenyl]carbonylpyrrolidin-3-yl]-3H-quinazolin-4-one, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-09 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of Pka and a Pkb Mimic.
Chemistry, 22, 2016
|
|
4UJ1
 
 | | Protein Kinase A in complex with an Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(3S,4R)-4-(3-chlorophenyl)carbonylpyrrolidin-3-yl]-3H-quinazolin-4-one, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of Pka and a Pkb Mimic.
Chemistry, 22, 2016
|
|
3A29
 
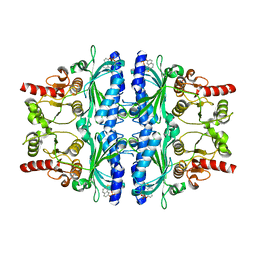 | | Crystal structure of human liver FBPase in complex with tricyclic inhibitor | | Descriptor: | 2-amino-4,5-dihydronaphtho[1,2-d][1,3]thiazol-8-yl dihydrogen phosphate, Fructose-1,6-bisphosphatase 1 | | Authors: | Takahashi, M, Sone, J, Hanzawa, H. | | Deposit date: | 2009-05-08 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis, SAR, and X-ray structure of tricyclic compounds as potent FBPase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7XDD
 
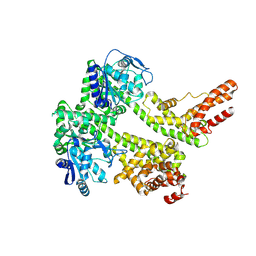 | | Cryo-EM structure of EDS1 and PAD4 | | Descriptor: | Lipase-like PAD4, Protein EDS1 | | Authors: | Huang, S.J, Jia, A.L, Sun, Y, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-03-26 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Science, 377, 2022
|
|
7P9T
 
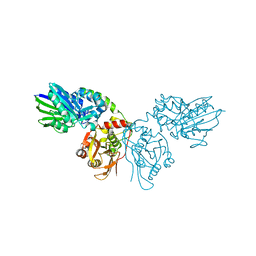 | | Crystal structure of CD73 in complex with dCMP in the open form | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
7PB5
 
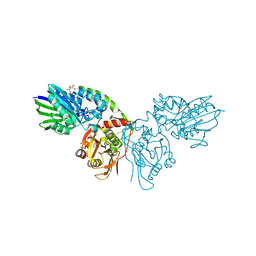 | | Crystal structure of CD73 in complex with UMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
