6WQ9
 
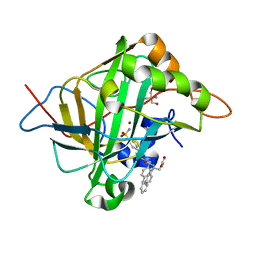 | | Carbonic Anhydrase II Complexed with 3-((2-((Naphthalen-2-ylmethyl)(4-sulfamoylphenethyl)amino)-2-oxoethyl)(phenethyl)amino)propanoic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
6WQ8
 
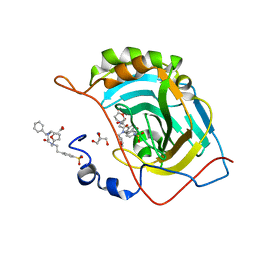 | | Carbonic Anhydrase II Complexed with 3-((2-((Furan-2-ylmethyl)(4-sulfamoylphenethyl)amino)-2-oxoethyl)(phenethyl)amino)propanoic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Combs, J.E, Lomelino, C, McKenna, R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | Sulfonamide Inhibitors of Human Carbonic Anhydrases Designed through a Three-Tails Approach: Improving Ligand/Isoform Matching and Selectivity of Action.
J.Med.Chem., 63, 2020
|
|
5MZU
 
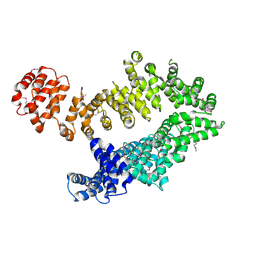 | |
3L2Y
 
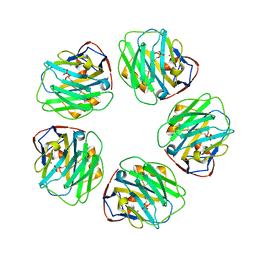 | | The structure of C-reactive protein bound to phosphoethanolamine | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | | Authors: | Mikolajek, H, Kolstoe, S.E, Wood, S.P, Pepys, M.B. | | Deposit date: | 2009-12-15 | | Release date: | 2010-12-08 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component.
J.Mol.Recognit., 24, 2011
|
|
1IL5
 
 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 2,5-DIAMINO-4,6-DIHYDROXYPYRIMIDINE (DDP) | | Descriptor: | 2,4-DIAMINO-4,6-DIHYDROXYPYRIMIDINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
4PVU
 
 | | Crystal structure of the complex between PPARgamma-LBD and the R enantiomer of Mbx-102 (Metaglidasen) | | Descriptor: | (2R)-(4-chlorophenyl)[3-(trifluoromethyl)phenoxy]ethanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Piemontese, L, Lavecchia, A. | | Deposit date: | 2014-03-18 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | On the metabolically active form of metaglidasen: improved synthesis and investigation of its peculiar activity on peroxisome proliferator-activated receptors and skeletal muscles.
Chemmedchem, 10, 2015
|
|
1V41
 
 | | Crystal structure of human PNP complexed with 8-Azaguanine | | Descriptor: | 5-AMINO-1H-[1,2,3]TRIAZOLO[4,5-D]PYRIMIDIN-7-OL, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Dos Santos, D.M, Canduri, F, Silva, R.G, Mendes, M.A, Basso, L.A, Palma, M.S, De Azevedo Jr, W.F, Santos, D.S. | | Deposit date: | 2003-11-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of human PNP complexed with ligands.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3OLS
 
 | | Crystal structure of estrogen receptor beta ligand binding domain | | Descriptor: | ESTRADIOL, Estrogen receptor beta, Nuclear receptor coactivator 1 | | Authors: | Moecklinghoff, S, Rose, R, Carraz, M, Visser, A, Ottmann, C, Brunsveld, L. | | Deposit date: | 2010-08-26 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and crystal structure of a phosphorylated estrogen receptor ligand binding domain.
Chembiochem, 11, 2010
|
|
2KZG
 
 | | A Transient and Low Populated Protein Folding Intermediate at Atomic Resolution | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Religa, T.L, Banachewicz, W, Fersht, A.R, Kay, L.E. | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A transient and low-populated protein-folding intermediate at atomic resolution.
Science, 329, 2010
|
|
5YSD
 
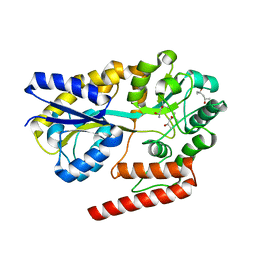 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
8FUG
 
 | |
3A11
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Thermococcus kodakaraensis KOD1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Translation initiation factor eIF-2B, ... | | Authors: | Nakamura, A, Fujihashi, M, Nishiba, Y, Yoshida, S, Yano, A, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-03-25 | | Release date: | 2010-03-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1
J.Biol.Chem., 287, 2012
|
|
1GZW
 
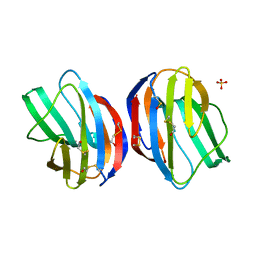 | | X-RAY CRYSTAL STRUCTURE OF HUMAN GALECTIN-1 | | Descriptor: | BETA-MERCAPTOETHANOL, GALECTIN-1, SULFATE ION, ... | | Authors: | Lopez-Lucendo, M.I.F, Gabius, H.J, Romero, A. | | Deposit date: | 2002-06-07 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Growth-Regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-Site Mutations and Their Impact on the Thermodynamics of Ligand Binding
J.Mol.Biol., 343, 2004
|
|
2DJQ
 
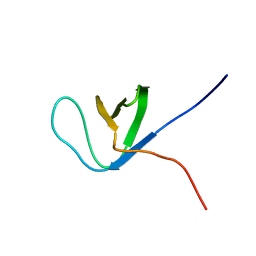 | | The solution structure of the first SH3 domain of mouse SH3 domain containing ring finger 2 | | Descriptor: | SH3 domain containing ring finger 2 | | Authors: | Sasagawa, A, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-05 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the first SH3 domain of mouse SH3 domain containing ring finger 2
To be Published
|
|
4IGY
 
 | | Crystal structure of kirola (Act d 11) - triclinic form | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
4IGV
 
 | | Crystal structure of kirola (Act d 11) | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
4IHR
 
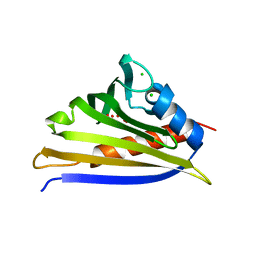 | | Crystal structure of recombinant kirola (Act d 11) | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Osinski, T, Majorek, K.A, Ciardiello, M.A, Chruszcz, M, Minor, W. | | Deposit date: | 2012-12-19 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
2L9V
 
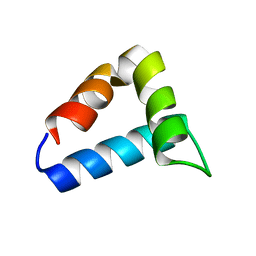 | | NMR structure of the FF domain L24A mutant's folding transition state | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Vernon, R.M, Religa, T.L, Hansen, A, Baker, D, Fersht, A.R, Kay, L.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: an NMR relaxation dispersion study.
J.Am.Chem.Soc., 133, 2011
|
|
5YSF
 
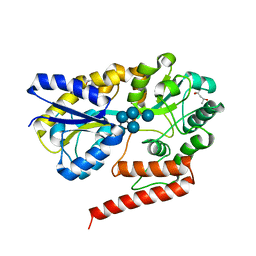 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
2QRW
 
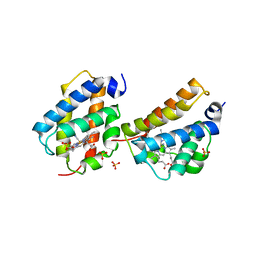 | | Crystal structure of Mycobacterium tuberculosis trHbO WG8F mutant | | Descriptor: | CYANIDE ION, Hemoglobin-like protein HbO, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Milani, M, Bolognesi, M. | | Deposit date: | 2007-07-30 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Roles of Tyr(CD1) and Trp(G8) in Mycobacterium tuberculosis Truncated Hemoglobin O in Ligand Binding and on the Heme Distal Site Architecture
Biochemistry, 46, 2007
|
|
8FW4
 
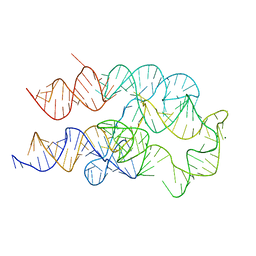 | |
4IGW
 
 | | Crystal structure of kirola (Act d 11) in P6122 space group | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Kirola, ... | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
3P2F
 
 | | Crystal structure of TofI in an apo form | | Descriptor: | AHL synthase | | Authors: | Yu, S, Rhee, S. | | Deposit date: | 2010-10-02 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibitor binding to an N-acyl-homoserine lactone synthase
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1N6U
 
 | | NMR structure of the interferon-binding ectodomain of the human interferon receptor | | Descriptor: | Interferon-alpha/beta receptor beta chain | | Authors: | Chill, J.H, Quadt, S.R, Levy, R, Schreiber, G, Anglister, J. | | Deposit date: | 2002-11-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The human type I interferon receptor. NMR structure reveals the molecular basis of ligand binding.
Structure, 11, 2003
|
|
6XTV
 
 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM WITH BOUND EFFECTOR ARG | | Descriptor: | ARGININE, Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
