1N51
 
 | | Aminopeptidase P in complex with the inhibitor apstatin | | Descriptor: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | Authors: | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-11-03 | | Release date: | 2003-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1BOA
 
 | | HUMAN METHIONINE AMINOPEPTIDASE 2 COMPLEXED WITH ANGIOGENESIS INHIBITOR FUMAGILLIN | | Descriptor: | COBALT (II) ION, FUMAGILLIN, METHIONINE AMINOPEPTIDASE | | Authors: | Liu, S, Widom, J, Kemp, C.W, Crews, C.M, Clardy, J.C. | | Deposit date: | 1998-08-01 | | Release date: | 1999-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
3H8F
 
 | | High pH native structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | BICARBONATE ION, Cytosol aminopeptidase, MANGANESE (II) ION, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
4K5O
 
 | |
1BPN
 
 | |
3IU8
 
 | | M. tuberculosis methionine aminopeptidase with Ni inhibitor T03 | | Descriptor: | 3-[(4-fluorobenzyl)sulfanyl]-4H-1,2,4-triazole, CHLORIDE ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2009-08-30 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalysis and Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidase
J.Med.Chem., 53, 2010
|
|
3IU9
 
 | | M. tuberculosis methionine aminopeptidase with Ni inhibitor T07 | | Descriptor: | 5-[(2,4-dichlorobenzyl)sulfanyl]-4H-1,2,4-triazol-3-amine, CHLORIDE ION, Methionine aminopeptidase, ... | | Authors: | Ye, Q.Z, Lu, J.P. | | Deposit date: | 2009-08-30 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalysis and Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidase
J.Med.Chem., 53, 2010
|
|
1BLL
 
 | |
1BPM
 
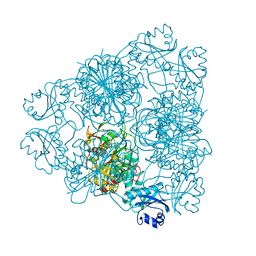 | |
1QXY
 
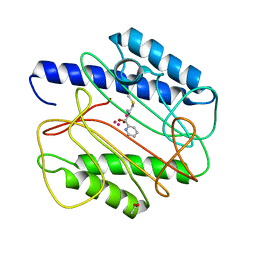 | | Crystal structure of S. aureus methionine aminopeptidase in complex with a ketoheterocycle 618 | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFANYL)-1-PYRIDIN-2-YLBUTANE-1,1-DIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
1QZY
 
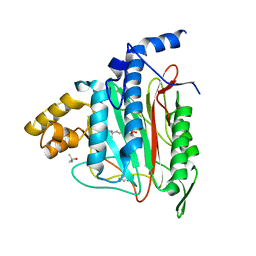 | | Human Methionine Aminopeptidase in complex with bengamide inhibitor LAF153 and cobalt | | Descriptor: | (E)-(2R,3R,4S,5R)-3,4,5-TRIHYDROXY-2-METHOXY-8,8-DIMETHYL-NON-6-ENOIC ACID ((3S,6R)-6-HYDROXY-2-OXO-AZEPAN-3-YL)-AMIDE, COBALT (II) ION, Methionine aminopeptidase 2, ... | | Authors: | Eck, M.J, Song, H.K, Morollo, A. | | Deposit date: | 2003-09-18 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proteomics-based target identification: bengamides as a new class of methionine aminopeptidase inhibitors.
J.Biol.Chem., 278, 2003
|
|
3OR3
 
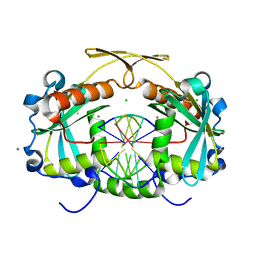 | | Restriction endonuclease HPY188I in complex with product DNA | | Descriptor: | 5'-D(*GP*AP*TP*CP*T)-3', 5'-D(*GP*TP*TP*CP*A)-3', 5'-D(P*GP*AP*AP*C)-3', ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2010-09-06 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hpy188I-DNA pre- and post-cleavage complexes--snapshots of the GIY-YIG nuclease mediated catalysis.
Nucleic Acids Res., 39, 2011
|
|
1QXZ
 
 | | Crystal structure of S. aureus methionine aminopeptidase in complex with a ketoheterocycle inhibitor 119 | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFANYL)-1-(1,3-THIAZOL-2-YL)BUTANE-1,1-DIOL, COBALT (II) ION, methionyl aminopeptidase | | Authors: | Douangamath, A, Dale, G.E, D'Arcy, A, Oefner, C. | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of staphylococcusaureus methionine aminopeptidase complexed with keto heterocycle and aminoketone inhibitors reveal the formation of a tetrahedral intermediate.
J.Med.Chem., 47, 2004
|
|
1R5H
 
 | | Crystal Structure of MetAP2 complexed with A320282 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-(2S,3R)-3-AMINO-4-CYCLOHEXYL-2-HYDROXY-BUTANO-N-(4-METHYLPHENYL)HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R58
 
 | | Crystal Structure of MetAP2 complexed with A357300 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-((2S,3R)-3-AMINO-2-HYDROXY-5-(ISOPROPYLSULFANYL)PENTANOYL)-N-3-CHLOROBENZOYL HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Ericken, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
5W2V
 
 | |
5VTT
 
 | | Dehaloperoxidase B Y38F mutant | | Descriptor: | Dehaloperoxidase B, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Carey, L.M, Ghiladi, R.A. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Probing the Structure-Function Relationship of a Multifunctional Enzyme using Crystallographic Diffraction Methods
Thesis, North Carolina State University, 2017
|
|
5W2D
 
 | |
5W30
 
 | | Crystal structure of mutant CJ YCEI protein (CJ-N48C) with monobromobimane guest structure | | Descriptor: | 3-(bromomethyl)-2,5,6-trimethyl-1H,7H-pyrazolo[1,2-a]pyrazole-1,7-dione, Putative periplasmic protein, SULFATE ION, ... | | Authors: | Huber, T.R, Snow, C.D. | | Deposit date: | 2017-06-07 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Installing Guest Molecules at Specific Sites within Scaffold Protein Crystals.
Bioconjug. Chem., 29, 2018
|
|
3PHJ
 
 | |
5W32
 
 | |
5W3C
 
 | |
3QHP
 
 | |
5VTS
 
 | | Dehaloperoxidase B Y28F mutant | | Descriptor: | Dehaloperoxidase B, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Carey, L.M, Ghiladi, R.A. | | Deposit date: | 2017-05-18 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Probing the Structure-Function Relationship of a Multifunctional Enzyme using Crystallographic Diffraction Methods
Thesis, North Carolina State University, 2017
|
|
5W2Z
 
 | |
