6DG1
 
 | |
2J69
 
 | | Bacterial dynamin-like protein BDLP | | Descriptor: | BACTERIAL DYNAMIN-LIKE PROTEIN | | Authors: | Low, H.H, Lowe, J. | | Deposit date: | 2006-09-26 | | Release date: | 2006-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Bacterial Dynamin-Like Protein
Nature, 444, 2006
|
|
7PY4
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P autoinhibited state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dieudonne, T, Abad-Herrera, S, Juknaviciute Laursen, M, Lejeune, M, Stock, C, Slimani, K, Jaxel, C, Lyons, J.A, Montigny, C, Gunther Pomorski, T, Nissen, P, Lenoir, G. | | Deposit date: | 2021-10-08 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Autoinhibition and regulation by phosphoinositides of ATP8B1, a human lipid flippase associated with intrahepatic cholestatic disorders.
Elife, 11, 2022
|
|
6E5E
 
 | |
6E5Q
 
 | |
6E62
 
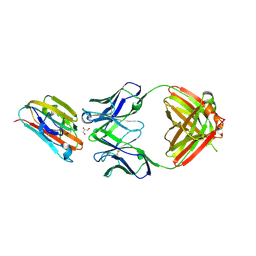 | | Crystal structure of malaria transmission-blocking antigen Pfs48/45 6C in complex with antibody 85RF45.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 85RF45.1 Fab heavy chain, 85RF45.1 Fab light chain, ... | | Authors: | Kundu, P, Semesi, A, Julien, J.P. | | Deposit date: | 2018-07-23 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural delineation of potent transmission-blocking epitope I on malaria antigen Pfs48/45.
Nat Commun, 9, 2018
|
|
6E5M
 
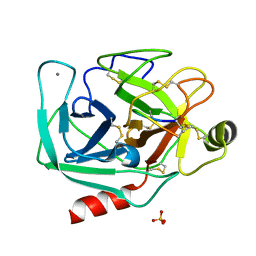 | | Crystallographic structure of the cyclic nonapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 32 2 1 | | Descriptor: | 9MER-PEPTIDE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Crystallographic structure of a complex between trypsin and a nonapeptide derived from a Bowman-Birk inhibitor found in Vigna unguiculata seeds.
Arch. Biochem. Biophys., 665, 2019
|
|
6E7M
 
 | |
2ZQZ
 
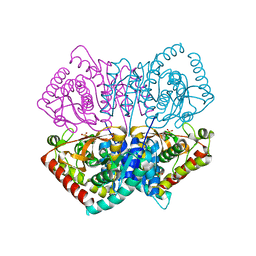 | | R-state structure of allosteric L-lactate dehydrogenase from Lactobacillus casei | | Descriptor: | L-lactate dehydrogenase, SULFATE ION | | Authors: | Arai, K, Ishimitsu, T, Fushinobu, S, Uchikoba, H, Matsuzawa, H, Taguchi, H. | | Deposit date: | 2008-08-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active and inactive state structures of unliganded Lactobacillus casei allosteric L-lactate dehydrogenase.
Proteins, 78, 2010
|
|
4POU
 
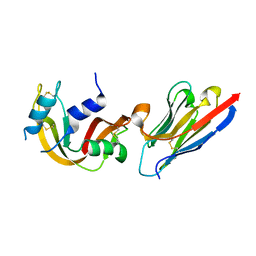 | | VHH-metal in Complex with RNase A | | Descriptor: | Ribonuclease pancreatic, VHH Antibody | | Authors: | Fanning, S.W, Walter, R, Horn, J.R. | | Deposit date: | 2014-02-26 | | Release date: | 2014-09-24 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of an engineered dual-specific antibody: conformational diversity leads to a hypervariable loop metal-binding site.
Protein Eng.Des.Sel., 27, 2014
|
|
6EGR
 
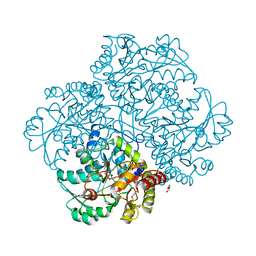 | | Crystal structure of Citrobacter freundii methionine gamma-lyase with V358Y replacement | | Descriptor: | DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Revtovich, S.V, Demitri, N, Raboni, S, Nikulin, A.D, Morozova, E.A, Demidkina, T.V, Storici, P, Mozzarelli, A. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering methionine gamma-lyase from Citrobacter freundii for anticancer activity.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
4PAS
 
 | | Heterodimeric coiled-coil structure of human GABA(B) receptor | | Descriptor: | Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Burmakina, S, Geng, Y, Chen, Y, Fan, Q.R. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Heterodimeric coiled-coil interactions of human GABAB receptor.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6E5S
 
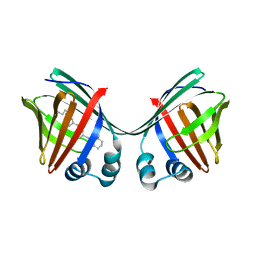 | |
6E65
 
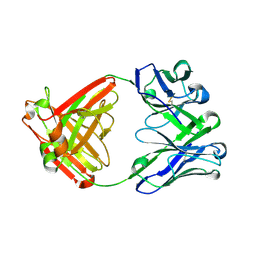 | |
4PU9
 
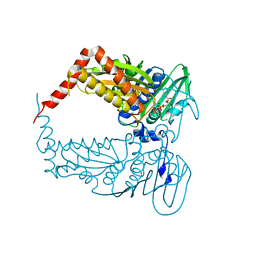 | |
1AUW
 
 | | H91N DELTA 2 CRYSTALLIN FROM DUCK | | Descriptor: | DELTA 2 CRYSTALLIN | | Authors: | Abu-Abed, M, Vallee, F, Howell, P.L. | | Deposit date: | 1997-09-03 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural comparison of the enzymatically active and inactive forms of delta crystallin and the role of histidine 91.
Biochemistry, 36, 1997
|
|
6E64
 
 | |
4PVL
 
 | | X-ray structure of human transthyretin (TTR) at room temperature to 1.9A resolution | | Descriptor: | Transthyretin | | Authors: | Fisher, S.J, Blakeley, M.P, Haupt, M, Mason, S.A, Cooper, J.B, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2014-03-18 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Binding site asymmetry in human transthyretin: insights from a joint neutron and X-ray crystallographic analysis using perdeuterated protein
IUCrJ, 1, 2014
|
|
5C9K
 
 | |
7PZB
 
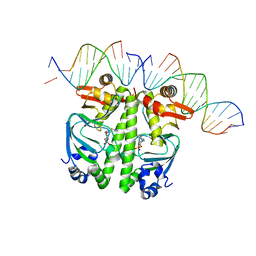 | | Structure of the Clr-cAMP-DNA complex | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*GP*TP*AP*AP*CP*AP*TP*TP*AP*CP*TP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*TP*AP*AP*TP*GP*TP*TP*AP*C)-3'), ... | | Authors: | Werel, L, Essen, L.-O. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Mbio, 14, 2023
|
|
7PZO
 
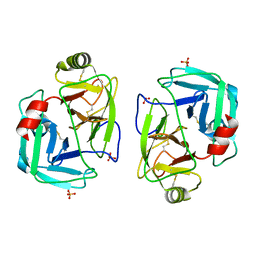 | | mite allergen Der p 3 from Dermatophagoides pteronyssinus | | Descriptor: | SULFATE ION, mite allergen Der p 3 | | Authors: | Timofeev, V.I, Shevtsov, M.B, Abramchik, Y.A, Mikheeva, O.O, Kostromina, M.A, Lykoshin, D.D, Zayats, E.A, Zavriev, S.K, Esipov, R.S, Kuranova, I.P. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural plasticity and thermal stability of the histone-like protein from Spiroplasma melliferum are due to phenylalanine insertions into the conservative scaffold.
J.Biomol.Struct.Dyn., 36, 2018
|
|
7PZA
 
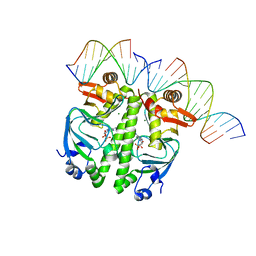 | | Structure of the Clr-cAMP-DNA complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*GP*TP*AP*AP*CP*AP*TP*TP*AP*CP*TP*CP*GP)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*TP*AP*AP*TP*GP*TP*TP*AP*C)-3'), ... | | Authors: | Werel, L, Essen, L.-O. | | Deposit date: | 2021-10-11 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Dual Specificity of Sinorhizobium meliloti Clr, a cAMP and cGMP Receptor Protein.
Mbio, 14, 2023
|
|
2J6P
 
 | | STRUCTURE OF AS-SB REDUCTASE FROM LEISHMANIA MAJOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SB(V)-AS(V) REDUCTASE, ... | | Authors: | Bisacchi, D, Zhou, Y, Rosen, B.P, Mukhopadhyay, R, Bordo, D. | | Deposit date: | 2006-10-02 | | Release date: | 2007-10-02 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of the As/Sb reductase LmACR2 from Leishmania major.
J. Mol. Biol., 386, 2009
|
|
7PSZ
 
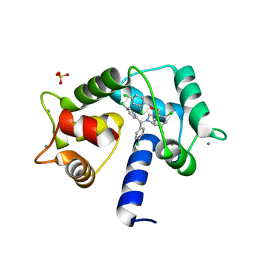 | | Crystal structure of CaM in complex with CDZ (form 1) | | Descriptor: | 1-[bis(4-chlorophenyl)methyl]-3-[(2~{R})-2-(2,4-dichlorophenyl)-2-[(2,4-dichlorophenyl)methoxy]ethyl]imidazole, CALCIUM ION, Calmodulin-1, ... | | Authors: | Mechaly, A.E, Leger, C, Haouz, A, Chenal, A. | | Deposit date: | 2021-09-24 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
Bmc Biol., 20, 2022
|
|
7PU9
 
 | | Crystal structure of CaM in complex with CDZ (form 2) | | Descriptor: | 1-[bis(4-chlorophenyl)methyl]-3-[(2~{R})-2-(2,4-dichlorophenyl)-2-[(2,4-dichlorophenyl)methoxy]ethyl]imidazole, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Leger, C, Haouz, A, Chenal, A. | | Deposit date: | 2021-09-28 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Dynamics and structural changes of calmodulin upon interaction with the antagonist calmidazolium.
Bmc Biol., 20, 2022
|
|
