7LWU
 
 | |
7LXW
 
 | |
7LY0
 
 | |
7LXZ
 
 | |
7LY2
 
 | |
7LXX
 
 | |
7LXY
 
 | |
7LY3
 
 | |
7LRT
 
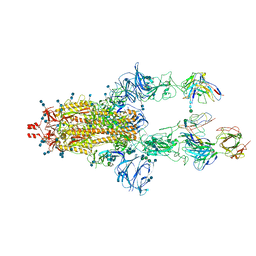 | |
7LRS
 
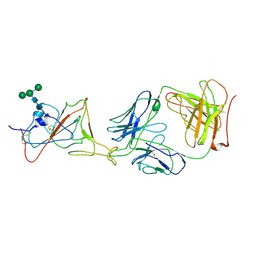 | |
7LC8
 
 | | SARS-CoV-2 spike Protein TM domain | | Descriptor: | Spike protein S2' | | Authors: | Fu, Q, Chou, J.J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Trimeric Hydrophobic Zipper Mediates the Intramembrane Assembly of SARS-CoV-2 Spike.
J.Am.Chem.Soc., 143, 2021
|
|
7M0J
 
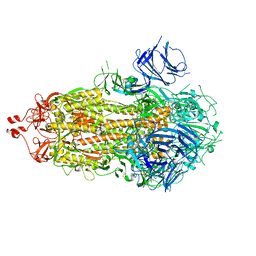 | |
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7M6F
 
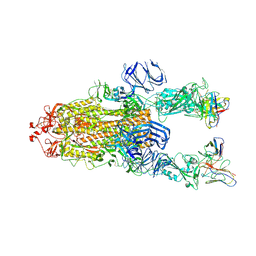 | |
7M6G
 
 | |
7M6I
 
 | |
7M8K
 
 | | Cryo-EM structure of Brazil (P.1) SARS-CoV-2 spike glycoprotein variant in the prefusion state (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Casner, R.G, Cerutti, G, Shapiro, L, Ho, D.D. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization.
Cell Host Microbe, 29, 2021
|
|
7M6E
 
 | |
7M6H
 
 | |
7M6D
 
 | |
7MJI
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 (focused refinement of RBD and VH ab8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab8 | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MKM
 
 | |
7M53
 
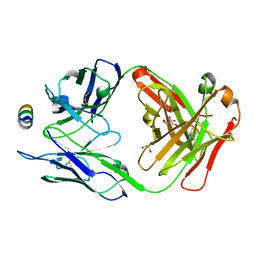 | | B6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | B6 antigen-binding (Fab) fragment heavy chain, B6 antigen-binding (Fab) fragment light chain, GLYCEROL, ... | | Authors: | Sauer, M.M, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for broad coronavirus neutralization.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7M7B
 
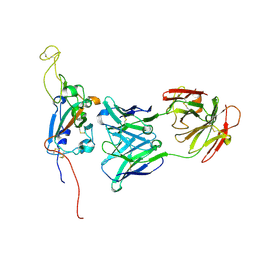 | | SARS-CoV-2 Spike:Fab 3D11 complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 3D11 heavy chain, Antibody Fab 3D11 light chain, ... | | Authors: | Asarnow, D, Cheng, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-05-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
7M71
 
 | | SARS-CoV-2 Spike:5A6 Fab complex I focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 5A6 Fab heavy chain, Antibody 5A6 Fab light chain, ... | | Authors: | Asarnow, D, Cheng, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
