9C9L
 
 | | Cryo-EM structure of the C1q A, B-crt, C peptide assembly narrow region | | Descriptor: | Peptide from Complement C1q subcomponent subunit A, Peptide from Complement C1q subcomponent subunit B, Peptide from Complement C1q subcomponent subunit C | | Authors: | Kreutzberger, M.A, Yu, L.T, Egelman, E.H, Hartgerink, J.D. | | Deposit date: | 2024-06-14 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A Collagen Triple Helix without the Superhelical Twist.
Acs Cent.Sci., 11, 2025
|
|
4IMV
 
 | |
4ZWI
 
 | | Surface Lysine Acetylated Human Carbonic Anhydrase II in Complex with a Sulfamate-Based Inhibitor | | Descriptor: | (6R)-1-O-acetyl-2,6-anhydro-6-{[4-(sulfamoyloxy)piperidin-1-yl]sulfonyl}-L-glucitol, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Mahon, B.P, McKenna, M. | | Deposit date: | 2015-05-19 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Observed surface lysine acetylation of human carbonic anhydrase II expressed in Escherichia coli.
Protein Sci., 24, 2015
|
|
3T7H
 
 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
7MG7
 
 | | Concanavalin A bound to a DNA glycoconjugate, Man-GTAC | | Descriptor: | 3-{[2-(2-hydroxyethoxy)ethyl]amino}-4-[(6-hydroxyhexyl)amino]cyclobut-3-ene-1,2-dione, CALCIUM ION, Concanavalin-A, ... | | Authors: | Partridge, B.E, Winegar, P.H, Mirkin, C.A. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Redefining Protein Interfaces within Protein Single Crystals with DNA.
J.Am.Chem.Soc., 143, 2021
|
|
9FGY
 
 | | Cryo-EM structure of Lysozyme homo-dimer assembled by homo Di-Gluebody - Local refinement | | Descriptor: | GLYCEROL, Lysozyme C, anti-Lysozyme Gluebody | | Authors: | Yi, G, Ye, M, Mamalis, D, Carrique, L, Fairhead, M, Li, H, Duerr, K, Zhang, P, Sauer, D.B, von Delft, F, Davis, B.G, Gilbert, R.J.C. | | Deposit date: | 2024-05-26 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Di-Gluebodies: Covalently-rigidified, modular protein assemblies enable simultaneous determination of high-resolution, low-size, cryo-EM structures
To Be Published
|
|
3SRP
 
 | |
7SCW
 
 | | KRAS full length wild-type in complex with RGL1 Ras association domain | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-09-29 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
1IB8
 
 | | SOLUTION STRUCTURE AND FUNCTION OF A CONSERVED PROTEIN SP14.3 ENCODED BY AN ESSENTIAL STREPTOCOCCUS PNEUMONIAE GENE | | Descriptor: | CONSERVED PROTEIN SP14.3 | | Authors: | Yu, L, Gunasekera, A.H, Mack, J, Olejniczak, E.T, Chovan, L.E, Ruan, X, Towne, D.L, Lerner, C.G, Fesik, S.W. | | Deposit date: | 2001-03-27 | | Release date: | 2002-03-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE AND FUNCTION OF A CONSERVED PROTEIN SP14.3 ENCODED BY AN ESSENTIAL STREPTOCOCCUS PNEUMONIAE GENE
J.Mol.Biol., 311, 2001
|
|
3SSX
 
 | | E. coli trp aporeporessor L75F mutant | | Descriptor: | TRIS(HYDROXYETHYL)AMINOMETHANE, Trp operon repressor | | Authors: | Benoff, B, Carey, J, Berman, H.M, Lawson, C.L. | | Deposit date: | 2011-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5801 Å) | | Cite: | Environment-dependent long-range structural distortion in a temperature-sensitive point mutant.
Protein Sci., 21, 2012
|
|
1HME
 
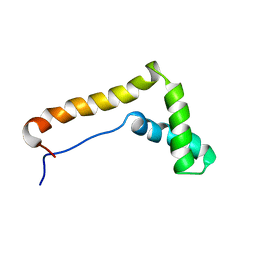 | | STRUCTURE OF THE HMG BOX MOTIF IN THE B-DOMAIN OF HMG1 | | Descriptor: | HIGH MOBILITY GROUP PROTEIN FRAGMENT-B | | Authors: | Weir, H.M, Kraulis, P.J, Hill, C.S, Raine, A.R.C, Laue, E.D, Thomas, J.O. | | Deposit date: | 1994-02-10 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the HMG box motif in the B-domain of HMG1.
EMBO J., 12, 1993
|
|
7YLQ
 
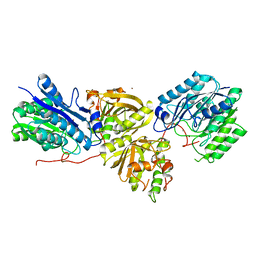 | | Crystal structure of Microcystinase C from Sphingomonas sp. ACM-3962 at 2.6 A resolution | | Descriptor: | CALCIUM ION, Microcystinase C, ZINC ION | | Authors: | Guo, X, Li, Z, Ding, W, Yin, P, Feng, L. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Microcystin C from Sphingomonas sp. ACM-3962 at 2.6 A resolution
To Be Published
|
|
9CF4
 
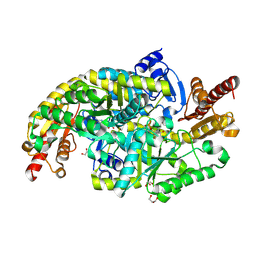 | | Crystal structure of the dimeric transaminase DoeD from C. salexigens DSM 3043. | | Descriptor: | Aminotransferase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Skogvold, A.C, Brakestad, H.T, Erlandsen, H, Leiros, I. | | Deposit date: | 2024-06-27 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Crystal structure and biochemical analysis of the dimeric transaminase DoeD provides insights into ectoine degradation.
Febs J., 292, 2025
|
|
7M3G
 
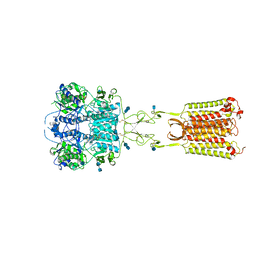 | | Asymmetric Activation of the Calcium Sensing Receptor Homodimer | | Descriptor: | 2-[4-[(3S)-3-[[(1R)-1-naphthalen-1-ylethyl]amino]pyrrolidin-1-yl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, Y, Robertson, M.J, Zhang, C, Meyerowitz, J.G, Panova, O, Skiniotis, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric activation of the calcium-sensing receptor homodimer.
Nature, 595, 2021
|
|
3V55
 
 | | Human MALT1 (334-719) in its ligand free form | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M, Wiesmann, C. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Determinants of MALT1 Protease Activity.
J.Mol.Biol., 419, 2012
|
|
7CSJ
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis-Complex with Coenzyme A and Gentamicin | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, COENZYME A, gentamicin C1 | | Authors: | Jeong, C.S, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Structural and biochemical analyses of an aminoglycoside 2'-N-acetyltransferase from Mycolicibacterium smegmatis.
Sci Rep, 10, 2020
|
|
1E0X
 
 | | XYLANASE 10A FROM SREPTOMYCES LIVIDANS. XYLOBIOSYL-ENZYME INTERMEDIATE AT 1.65 A | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL, beta-D-xylopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-xylopyranose | | Authors: | Ducros, V, Charnock, S.J, Derewenda, U, Derewenda, Z.S, Dauter, Z, Dupont, C, Shareck, F, Morosoli, R, Kluepfel, D, Davies, G.J. | | Deposit date: | 2000-04-10 | | Release date: | 2001-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Specificity in Glycoside Hydrolase Family 10. Structural and Kinetic Analysis of the Streptomyces Lividans Xylanase 10A
J.Biol.Chem., 275, 2000
|
|
4ZUO
 
 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a hydroxamate inhibitor | | Descriptor: | 6-[(3-aminopropyl)amino]-N-hydroxyhexanamide, AMMONIUM ION, Acetylpolyamine aminohydrolase, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2015-05-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
3AFF
 
 | | Crystal structure of the HsaA monooxygenase from M. tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Strynadka, N, Eltis, L.D. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
4IMB
 
 | |
2WN8
 
 | |
8XXT
 
 | | ASFV RNAP M1249L C-tail occupied complex2 (MCOC2) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
382D
 
 | |
2L8O
 
 | | Solution structure of Chr148 from Cytophaga hutchinsonii, Northeast Structural Genomics Consortium Target Chr148 | | Descriptor: | Activator of Hsp90 ATPase homologue 1-like C-terminal domain-containing protein | | Authors: | Liu, Y, Lee, D, Ciccosanti, C, Nair, L, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-01-21 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Chr148 from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target Chr148
To be Published
|
|
2WN4
 
 | |
