6QM3
 
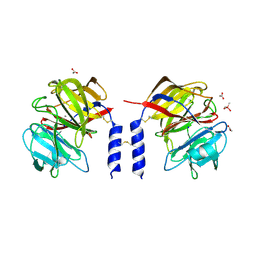 | | Crystal structure of a calcium- and sodium-bound mouse Olfactomedin-1 disulfide-linked dimer of the Olfactomedin domain and part of coiled coil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CACODYLATE ION, ... | | Authors: | Pronker, M.F, van den Hoek, H.G, Janssen, B.J.C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
BMC Mol Cell Biol, 20, 2019
|
|
3DNI
 
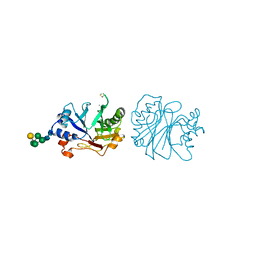 | | CRYSTALLOGRAPHIC REFINEMENT AND STRUCTURE OF DNASE I AT 2 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, DEOXYRIBONUCLEASE I, alpha-D-galactopyranose-(1-6)-beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oefner, C, Suck, D. | | Deposit date: | 1992-08-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic refinement and structure of DNase I at 2 A resolution.
J.Mol.Biol., 192, 1986
|
|
8F9X
 
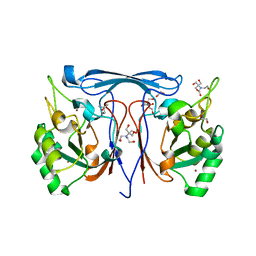 | | Cyclase-Phosphotriesterase from Ruegeria pomeroyi DSS-3 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Frkic, R.L, Ji, D, Jackson, C.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-06 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A Thermostable Bacterial Metallohydrolase that Degrades Organophosphate Plasticizers.
Chembiochem, 26, 2025
|
|
7KFA
 
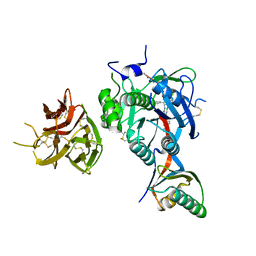 | | PCSK9 in complex with PCSK9i a 13mer cyclic peptide LDLR disruptor | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Chopra, R, Xu, M, Spraggon, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
6U1P
 
 | | Crystal structure of VpsU (VC0916) from Vibrio cholerae | | Descriptor: | GLYCEROL, Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Tripathi, S.M, Schwechheimer, C, Herbert, K, Osorio, J, Yildiz, F.H, Rubin, S.M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | A tyrosine phosphoregulatory system controls exopolysaccharide biosynthesis and biofilm formation in Vibrio cholerae.
Plos Pathog., 16, 2020
|
|
6IWR
 
 | | Crystal structure of GalNAc-T7 with UDP, GalNAc and Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7, ... | | Authors: | Yu, C, Yin, Y.X. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
1IU2
 
 | | The first PDZ domain of PSD-95 | | Descriptor: | PSD-95 | | Authors: | Long, J.-F, Tochio, H, Wang, P, Sala, C, Niethammer, M, Sheng, M, Zhang, M. | | Deposit date: | 2002-02-19 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95
J.MOL.BIOL., 327, 2003
|
|
2K9C
 
 | | Paramagnetic shifts in solid-state NMR of Proteins to elicit structural information | | Descriptor: | COBALT (II) ION, Macrophage metalloelastase | | Authors: | Balayssac, S, Bertini, I, Bhaumik, A, Lelli, M, Luchinat, C. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Paramagnetic shifts in solid-state NMR of proteins to elicit structural information
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2K6D
 
 | | CIN85 Sh3-C domain in complex with ubiquitin | | Descriptor: | SH3 domain-containing kinase-binding protein 1, Ubiquitin | | Authors: | Forman-Kay, J, Bezsonova, I. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interactions between the three CIN85 SH3 domains and ubiquitin: implications for CIN85 ubiquitination.
Biochemistry, 47, 2008
|
|
8DS0
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp14-nsp15 (C14) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS2
 
 | | Structure of SARS-CoV-2 Mpro in complex with the nsp13-nsp14 (C13) cut site sequence (form 2) | | Descriptor: | 3C-like proteinase nsp5, GLYCEROL, SODIUM ION | | Authors: | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
2W43
 
 | | Structure of L-haloacid dehalogenase from S. tokodaii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HYPOTHETICAL 2-HALOALKANOIC ACID DEHALOGENASE, PHOSPHATE ION | | Authors: | Rye, C.A, Isupov, M.N, Lebedev, A.A, Littlechild, J.A. | | Deposit date: | 2008-11-21 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Biochemical and Structural Studies of a L-Haloacid Dehalogenase from the Thermophilic Archaeon Sulfolobus Tokodaii.
Extremophiles, 13, 2009
|
|
6UG6
 
 | | C3 symmetric peptide design number 1, Sporty, crystal form 2 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, C3-1, ... | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-25 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
1DL4
 
 | | THE SOLUTION STRUCTURE OF A BAY-REGION 1S-BENZ[A]ANTHRACENE OXIDE ADDUCT AT THE N6 POSITION OF ADENINE OF AN OLIGODEOXYNUCLEOTIDE CONTAINING THE HUMAN N-RAS CODON 61 SEQUENCE | | Descriptor: | 1R,2S,3R,4S-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, DNA (5'-D(*CP*GP*GP*AP*CP*(BZA)AP*AP*GP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3') | | Authors: | Li, Z, Kim, H.-Y, Tamura, P.J, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 1999-12-08 | | Release date: | 2000-01-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Intercalation of the (1S,2R,3S,4R)-N6-[1-(1,2,3,4-tetrahydro-2,3, 4-trihydroxybenz[a]anthracenyl)]-2'-deoxyadenosyl adduct in an oligodeoxynucleotide containing the human N-ras codon 61 sequence.
Biochemistry, 38, 1999
|
|
8XE8
 
 | | Solution structure of ubiquitin-like domain (UBL) of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1 | | Authors: | Lai, C.H, Ko, K.T, Fan, P.J, Yu, T.A, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2023-12-11 | | Release date: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the ZFAND1-p97 interaction involved in stress granule clearance.
J.Biol.Chem., 300, 2024
|
|
2ONT
 
 | | A swapped dimer of the HIV-1 capsid C-terminal domain | | Descriptor: | Capsid protein p24 | | Authors: | Ivanov, D, Tsodikov, O.V, Kasanov, J, Ellenberger, T, Wagner, G, Collins, T. | | Deposit date: | 2007-01-24 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Domain-swapped dimerization of the HIV-1 capsid C-terminal domain
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
9B5Y
 
 | | Cryo-EM structure of the LAPTH-bound PTH1R in complex with Gq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q), ... | | Authors: | Zhang, X, Liu, H, Zhang, C, Vilardaga, J.-P. | | Deposit date: | 2024-03-22 | | Release date: | 2025-03-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Allosteric mechanism in the distinctive coupling of G q and G s to the parathyroid hormone type 1 receptor.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
4XF2
 
 | | Tetragonal structure of Arp2/3 complex | | Descriptor: | Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, Actin-related protein 2/3 complex subunit 2, ... | | Authors: | Jurgenson, C.T, Pollard, T.P. | | Deposit date: | 2014-12-25 | | Release date: | 2015-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Crystals of the Arp2/3 complex in two new space groups with structural information about actin-related protein 2 and potential WASP binding sites.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4I5Q
 
 | | Crystal structure and catalytic mechanism for peroplasmic disulfide-bond isomerase DsbC from Salmonella enterica serovar Typhimurium | | Descriptor: | MAGNESIUM ION, Thiol:disulfide interchange protein DsbC | | Authors: | Ha, N.C, Li, J, Kim, J.S, Yoon, B.Y, Yeom, J.H, Lee, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Crystal structure of the periplasmic disulfide-bond isomerase DsbC from Salmonella enterica serovar Typhimurium and the mechanistic implications.
J.Struct.Biol., 183, 2013
|
|
8XA0
 
 | | penton capsomer of the VZV C-capsid | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Nan, W, Lei, C, Xiangxi, W. | | Deposit date: | 2023-12-01 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Hlife, 2, 2024
|
|
6TY5
 
 | | Crystal structure of human TLR8 in complex with Compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-7-(7-methyl-2-piperidin-4-yl-indazol-5-yl)furo[3,2-c]pyridin-4-one, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Target-Based Identification and Optimization of 5-Indazol-5-yl Pyridones as Toll-like Receptor 7 and 8 Antagonists Using a Biochemical TLR8 Antagonist Competition Assay.
J.Med.Chem., 63, 2020
|
|
1DN9
 
 | |
2WQO
 
 | | STRUCTURE OF NEK2 BOUND TO THE AMINOPYRIDINE CCT241950 | | Descriptor: | 4-[2-AMINO-5-(3,4,5-TRIMETHOXYPHENYL)PYRIDIN-3-YL]BENZOIC ACID, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2009-08-24 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.167 Å) | | Cite: | An Autoinhibitory Tyrosine Motif in the Cell-Cycle- Regulated Nek7 Kinase is Released Through Binding of Nek9.
Mol.Cell, 36, 2009
|
|
8PE1
 
 | | Crystal structure of Gel4 in complex with Nanobody 4 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 4, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
2X65
 
 | | Crystal structure of T. maritima GDP-mannose pyrophosphorylase in complex with mannose-1-phosphate. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-O-phosphono-alpha-D-mannopyranose, ... | | Authors: | Pelissier, M.C, Lesley, S, Kuhn, P, Bourne, Y. | | Deposit date: | 2010-02-15 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights Into the Catalytic Mechanism of Bacterial Guanosine-Diphospho-D-Mannose Pyrophosphorylase and its Regulation by Divalent Ions.
J.Biol.Chem., 285, 2010
|
|
