1LPK
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 125. | | Descriptor: | 1-(3-CARBAMIMIDOYL-BENZYL)-1H-INDOLE-2-CARBOXYLIC ACID 3-CARBAMIMIDOYL-BENZYLESTER, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Loenze, P, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Med.Chem., 45, 2002
|
|
6H51
 
 | | Crystal structure of human KDM5B in complex with compound 34f | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-[1-(cyclobutylmethyl)piperidin-4-yl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
1LQD
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 45. | | Descriptor: | 1-(3-CARBAMIMIDOYL-BENZYL)-4-METHYL-1H-INDOLE-2-CARBOXYLIC ACID 3,5-DIMETHYL-BENZYLAMIDE, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Loenze, P, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-10 | | Release date: | 2003-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Med.Chem., 45, 2002
|
|
7MSG
 
 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
1LPG
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 79. | | Descriptor: | Blood coagulation factor Xa, CALCIUM ION, [4-({[5-BENZYLOXY-1-(3-CARBAMIMIDOYL-BENZYL)-1H-INDOLE-2-CARBONYL]-AMINO}-METHYL)-PHENYL]-TRIMETHYL-AMMONIUM | | Authors: | Schreuder, H.A, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa.
J.Med.Chem., 45, 2002
|
|
3C45
 
 | | Human dipeptidyl peptidase IV/CD26 in complex with a fluoroolefin inhibitor | | Descriptor: | (2S,3S)-3-{3-[2-chloro-4-(methylsulfonyl)phenyl]-1,2,4-oxadiazol-5-yl}-1-cyclopentylidene-4-cyclopropyl-1-fluorobutan-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Edmondson, S.D, Weber, A.E. | | Deposit date: | 2008-01-29 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Fluoroolefins as amide bond mimics in dipeptidyl peptidase IV inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1VJY
 
 | | Crystal Structure of a Naphthyridine Inhibitor of Human TGF-beta Type I Receptor | | Descriptor: | 2-[5-(6-METHYLPYRIDIN-2-YL)-2,3-DIHYDRO-1H-PYRAZOL-4-YL]-1,5-NAPHTHYRIDINE, TGF-beta receptor type I | | Authors: | Gellibert, F, Woolven, J, Fouchet, M.-H, Mathews, N, Goodland, H, Lovegrove, V, Laroze, A, Nguyen, V.-L, Sautet, S, Wang, R, Janson, C, Smith, W, Krysa, G, Boullay, V, de Gouville, A.-C, Huet, S, Hartley, D. | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of 1,5-Naphthyridine Derivatives as a Novel Series of Potent and Selective TGF-beta Type I Receptor Inhibitors.
J.Med.Chem., 47, 2004
|
|
4UZD
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[3-(1H-benzimidazol-2-yloxy)phenyl]-8-oxo-4,5,6,7,8,9-hexahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Pouzieux, S, Delarbre, L, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
8F45
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a phenyl dimethyl sulfane inhibitor (cyclopropyl ketoamide warhead) | | Descriptor: | (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(2~{S},3~{S})-3-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
8F46
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl phenyl sulfane inhibitor (cyano warhead) | | Descriptor: | 3C-like proteinase, N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucinamide, TETRAETHYLENE GLYCOL | | Authors: | Liu, L, Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
2GHL
 
 | | Mutant Mus Musculus P38 Kinase Domain in Complex with Inhibitor PG-874743 | | Descriptor: | 3-(2-CHLOROPHENYL)-1-(2-{[(1S)-2-HYDROXY-1,2-DIMETHYLPROPYL]AMINO}PYRIMIDIN-4-YL)-1-(4-METHOXYPHENYL)UREA, Mitogen-activated protein kinase 14 | | Authors: | Walter, R.L, Mekel, M.J, Evdokimov, A.G, Pokross, M.E, Brugel, T.A. | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Development of N-2,4-pyrimidine-N-phenyl-N'-phenyl ureas as inhibitors of tumor necrosis factor alpha (TNF-alpha) synthesis. Part 1.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
8F44
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a dimethyl phenyl sulfane inhibitor | | Descriptor: | (1R,2S)-1-hydroxy-2-[(N-{[2-methyl-2-(phenylsulfanyl)propoxy]carbonyl}-L-leucyl)amino]-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (2-methyl-2-phenylsulfanyl-propyl) ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase, ... | | Authors: | Liu, L, Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2022-11-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV).
Eur.J.Med.Chem., 254, 2023
|
|
6OVA
 
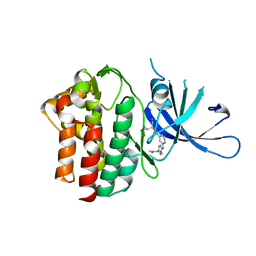 | | Crystal Structure of TYK2 with novel pyrrolidinone inhibitor | | Descriptor: | 6-({4-[(3S)-3-cyano-3-cyclopropyl-2-oxopyrrolidin-1-yl]pyridin-2-yl}amino)-N,N-dimethylpyridine-3-carboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Skene, R.J, Hoffman, I.D. | | Deposit date: | 2019-05-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Efficient synthesis of tert-butyl 3-cyano-3-cyclopropyl-2-oxopyrrolidine-4-carboxylates: Highly functionalized 2-pyrrolidinone enabling access to novel macrocyclic Tyk2 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8HJE
 
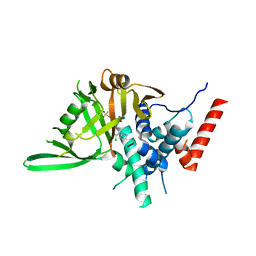 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
3GMZ
 
 | |
4UZH
 
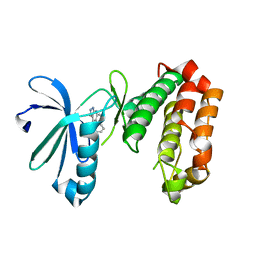 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | (4S)-4-(2-fluorophenyl)-2,4,6,7,8,9-hexahydro-5H-pyrazolo[3,4-b][1,7]naphthyridin-5-one, AURORA 2 KINASE DOMAIN | | Authors: | Pouzieux, S, Maignan, S, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
2ZAZ
 
 | | Crystal structure of P38 in complex with 4-anilino quinoline inhibitor | | Descriptor: | 4-{4-[(5-hydroxy-2-methylphenyl)amino]quinolin-7-yl}-1,3-thiazole-2-carbaldehyde, ACETATE ION, GLYCEROL, ... | | Authors: | Somers, D.O. | | Deposit date: | 2007-10-12 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biphenyl amide p38 kinase inhibitors 1: Discovery and binding mode
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6H4T
 
 | | Crystal structure of human KDM4A in complex with compound 19a | | Descriptor: | 8-[4-(2-spiro[1,2-dihydroindene-3,4'-piperidine]-1'-ylethyl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6H4O
 
 | | Crystal structure of human KDM4A in complex with compound 18a | | Descriptor: | 8-[4-[2-[4-[3-(trifluoromethyl)phenyl]piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
8FUR
 
 | | Crystal structure of human IDO1 with compound 11 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(4-methylphenyl)-N'-[(1P,2'P)-4-propoxy-5-propyl-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-yl]urea | | Authors: | Critton, D.A, Lewis, H.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Synthesis and biological evaluation of biaryl alkyl ethers as inhibitors of IDO1.
Bioorg.Med.Chem.Lett., 88, 2023
|
|
6H4Q
 
 | | Crystal structure of human KDM4A in complex with compound 34a | | Descriptor: | 8-[4-(1-methylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6H4W
 
 | | Crystal structure of human KDM4A in complex with compound 19d | | Descriptor: | 8-[4-[2-[4-(3-chlorophenyl)-4-methyl-piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
8FIV
 
 | |
8FIW
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor Jun10221 | | Descriptor: | 3C-like proteinase nsp5, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide, N-([1,1'-biphenyl]-4-yl)-N-[(1S)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]prop-2-enamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Exploring diverse reactive warheads for the design of SARS-CoV-2 main protease inhibitors.
Eur.J.Med.Chem., 259, 2023
|
|
7QDN
 
 | |
