4ZCG
 
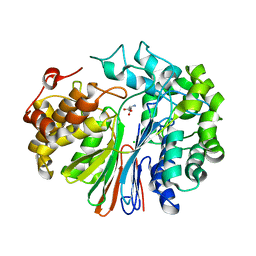 | | Crystal Structure of human GGT1 in complex with Glutamate (with all atoms of glutamate) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Terzyan, S, Hanigan, M. | | Deposit date: | 2015-04-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Human gamma-Glutamyl Transpeptidase 1: STRUCTURES OF THE FREE ENZYME, INHIBITOR-BOUND TETRAHEDRAL TRANSITION STATES, AND GLUTAMATE-BOUND ENZYME REVEAL NOVEL MOVEMENT WITHIN THE ACTIVE SITE DURING CATALYSIS.
J.Biol.Chem., 290, 2015
|
|
2ZJI
 
 | |
4TYU
 
 | |
6HMX
 
 | |
4TXY
 
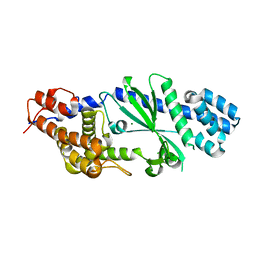 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase, a prokaryotic cGAS homolog | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.0001 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
6V10
 
 | | genome-containing AAV8 particles | | Descriptor: | Capsid protein, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
5JMR
 
 | | X-ray structure of the furin inhibitory antibody Nb14 | | Descriptor: | CHLORIDE ION, camelid VHH fragment | | Authors: | Dahms, S.O, Than, M.E. | | Deposit date: | 2016-04-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers.
Sci Rep, 6, 2016
|
|
1NR8
 
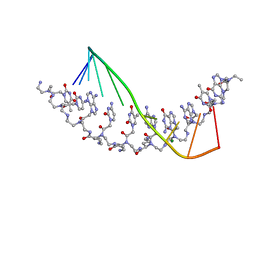 | | The crystal structure of a D-Lysine-based chiral PNA-DNA duplex | | Descriptor: | 5'-D(P*AP*GP*TP*GP*AP*TP*CP*TP*AP*C)-3', H-((GPN)*(TPN)*(APN)*(GPN)*(A66)*(T66)*(C66)*(APN)*(CPN)*(TPN))-NH2, MAGNESIUM ION | | Authors: | Menchise, V, De Simone, G, Tedeschi, T, Corradini, R, Sforza, S, Marchelli, R, Capasso, D, Saviano, M, Pedone, C. | | Deposit date: | 2003-01-24 | | Release date: | 2003-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into peptide nucleic acid (PNA) structural features: The
crystal structure of a D-lysine-based chiral PNA-DNA duplex
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7RZ8
 
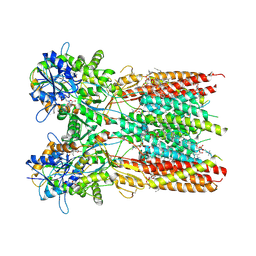 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RZA
 
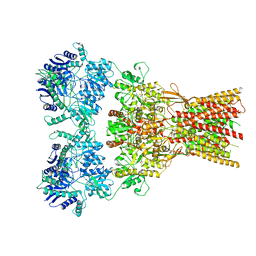 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RZ6
 
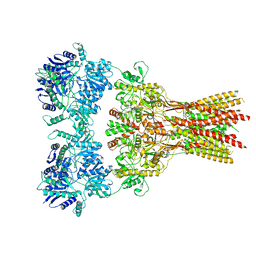 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist glutamate | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RYZ
 
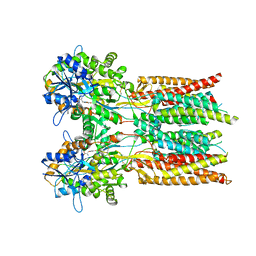 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
4U7S
 
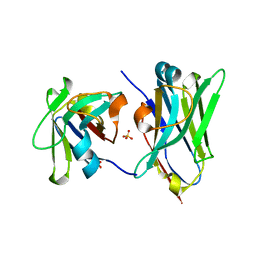 | |
7RYY
 
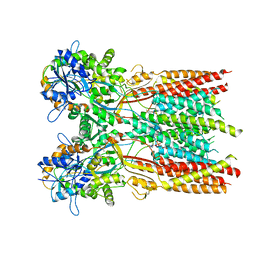 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist glutamate | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTAMIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RZ5
 
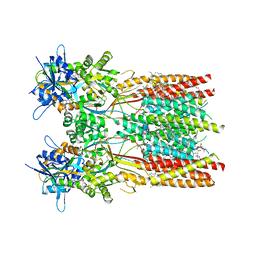 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK 200775 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
7RZ9
 
 | |
7RZ4
 
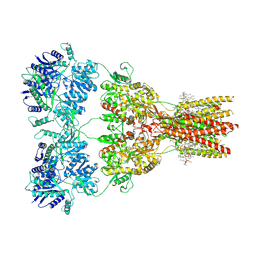 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK 200775 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, ... | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
1JQW
 
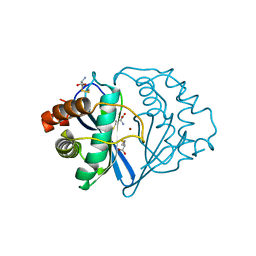 | | THE 2.3 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/HOMOCYSTEINE COMPLEX | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-09 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
7RZ7
 
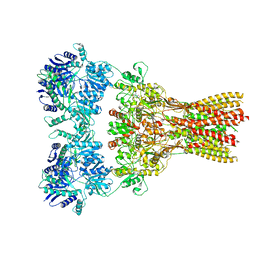 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to agonist Quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Klykov, O.V, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
4TZP
 
 | | As Grown, Untreated Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its cognate tRNAGly, and B. subtilis YbxF protein | | Descriptor: | Ribosome-associated protein L7Ae-like, engineered tRNA, glyQ T-box Stem I | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-10 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (8.503 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
4TZZ
 
 | | Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its Cognate tRNAGly, and B. subtilis YbxF protein, treated by removing lithium sulfate and increasing PEG3350 concentration from 20% to 45% post crystallization | | Descriptor: | MAGNESIUM ION, Ribosome-associated protein L7Ae-like, T-box Stem I RNA, ... | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
1JVI
 
 | | THE 2.2 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/RIBOSILHOMOCYSTEINE COMPLEX | | Descriptor: | (2S)-2-amino-4-[[(2S,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methylsulfanyl]butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ... | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
6V1G
 
 | | Genome-containing AAVrh.10 | | Descriptor: | Capsid protein VP1, DNA (5'-D(*CP*A)-3') | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
2VOE
 
 | | Crystal structure of Rv2780 from M. tuberculosis H37Rv | | Descriptor: | ALANINE DEHYDROGENASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Mycobacterium Tuberculosis Secretory Antigen Alanine Dehydrogenase (Rv2780) in Apo and Ternary Complex Forms Captures "Open" and "Closed" Enzyme Conformations.
Proteins, 72, 2008
|
|
6TKV
 
 | | Crystal structure of the human FUT8 in complex with GDP and a biantennary complex N-glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-(1,6)-fucosyltransferase, GLYCEROL, ... | | Authors: | Garcia-Garcia, A, Ceballos-Laita, L, Serna, L, Artschwager, R, Reichardt, N.C, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2019-11-29 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate specificity and catalysis of alpha 1,6-fucosyltransferase.
Nat Commun, 11, 2020
|
|
