8U5Y
 
 | | human RADX trimer bound to ssDNA | | Descriptor: | DNA (25-MER), RPA-related protein RADX | | Authors: | Balakrishnan, S, Chazin, W.J. | | Deposit date: | 2023-09-13 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structure of RADX and mechanism for regulation of RAD51 nucleofilaments.
Biorxiv, 2023
|
|
5Z51
 
 | |
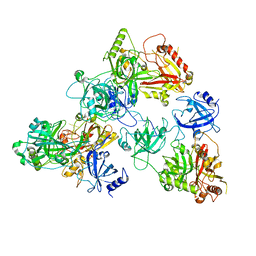
2XCQ
 
 | | The 2.98A crystal structure of the catalytic core (B'A' region) of Staphylococcus aureus DNA Gyrase | | Descriptor: | DNA GYRASE SUBUNIT B, DNA GYRASE SUBUNIT A | | Authors: | Bax, B.D, Chan, P.F, Eggleston, D.S, Fosberry, A, Gentry, D.R, Gorrec, F, Giordano, I, Hann, M.M, Hennessy, A, Hibbs, M, Huang, J, Jones, E, Jones, J, Brown, K.K, Lewis, C.J, May, E.W, Singh, O, Spitzfaden, C, Shen, C, Shillings, A, Theobald, A.F, Wohlkonig, A, Pearson, N.D, Gwynn, M.N. | | Deposit date: | 2010-04-24 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Type Iia Topoisomerase Inhibition by a New Class of Antibacterial Agents.
Nature, 466, 2010
|
|
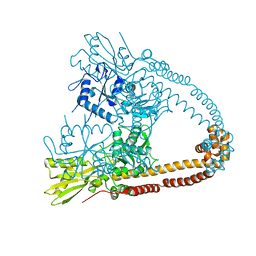
5WW7
 
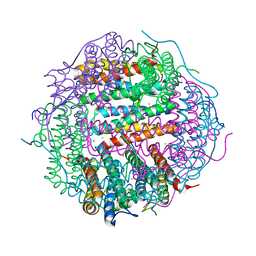 | |
5WW6
 
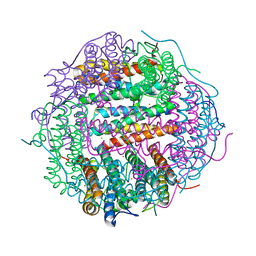 | |
5WW5
 
 | |
5WW8
 
 | |
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
5WW4
 
 | |
5WW3
 
 | |
5WW9
 
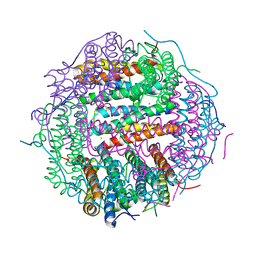 | |
7EUC
 
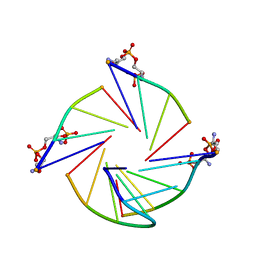 | | Parallel G-quadruplex structure | | Descriptor: | DNA (5'-D(*TP*TP*GP*GP*GP*G)-3'), DNA (5'-D(P*GP*GP*GP*G)-3'), DNA (5'-D(P*GP*GP*GP*GP*T)-3'), ... | | Authors: | Winnerdy, F.R, Devi, G, Ang, J.C.Y, Lim, K.W, Phan, A.T. | | Deposit date: | 2021-05-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Four-Layered Intramolecular Parallel G-Quadruplex with Non-Nucleotide Loops: An Ultra-Stable Self-Folded DNA Nano-Scaffold.
Acs Nano, 16, 2022
|
|
7N8S
 
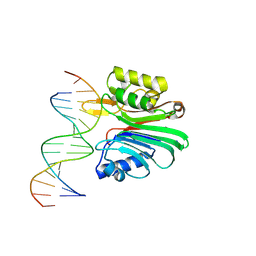 | | LINE-1 endonuclease domain complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*TP*AP*AP*AP*AP*AP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*TP*CP*CP*TP*TP*TP*TP*TP*AP*AP*GP*GP*A)-3'), LINE-1 retrotransposable element ORF2 protein | | Authors: | Korolev, S, Miller, I. | | Deposit date: | 2021-06-15 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural dissection of sequence recognition and catalytic mechanism of human LINE-1 endonuclease.
Nucleic Acids Res., 49, 2021
|
|
7N94
 
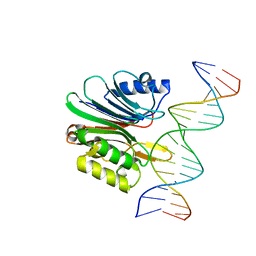 | | LINE-1 endonuclease domain complex with DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*CP*TP*TP*AP*AP*AP*AP*AP*GP*GP*AP*GP*CP*T)-3'), DNA (5'-D(*GP*CP*TP*CP*CP*TP*TP*TP*TP*TP*AP*AP*GP*GP*GP*CP*TP*A)-3'), LINE-1 retrotransposable element ORF2 protein | | Authors: | Korolev, S, Miller, I. | | Deposit date: | 2021-06-16 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural dissection of sequence recognition and catalytic mechanism of human LINE-1 endonuclease.
Nucleic Acids Res., 49, 2021
|
|
5FPR
 
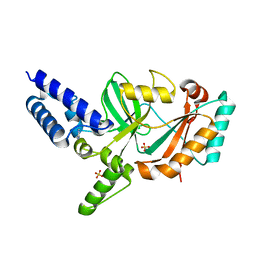 | | Structure of Bacterial DNA Ligase with small-molecule ligand pyrimidin-2-amine (AT371) in an alternate binding site. | | Descriptor: | DNA LIGASE, PYRIMIDIN-2-AMINE, SULFATE ION | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2QNF
 
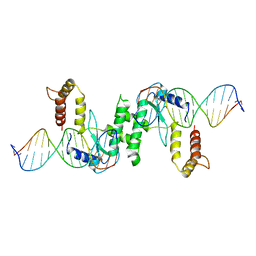 | | Crystal structure of T4 Endonuclease VII H43N mutant in complex with heteroduplex DNA containing base mismatches | | Descriptor: | DNA (5'-D(*DCP*DAP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DGP*DAP*DGP*DCP*DCP*DG)-3'), DNA (5'-D(*DCP*DAP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DGP*DAP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(*DCP*DGP*DGP*DCP*DTP*DCP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DTP*DG)-3'), ... | | Authors: | Biertumpfel, C, Yang, W, Suck, D. | | Deposit date: | 2007-07-18 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of T4 endonuclease VII resolving a Holliday junction.
Nature, 449, 2007
|
|
2IF9
 
 | |
8C7B
 
 | |
5ZOG
 
 | | Crystal Structure of R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
8XJ8
 
 | | The Cryo-EM structure of MPXV E5 C-terminal in complex with DNA | | Descriptor: | DNA (70-MER), MAGNESIUM ION, Monkeypox virus E5, ... | | Authors: | Zhang, W, Liu, Y, Gao, H, Gan, J. | | Deposit date: | 2023-12-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
8XJ7
 
 | | The Cryo-EM structure of MPXV E5 in complex with DNA | | Descriptor: | DNA (70-MER), MAGNESIUM ION, Monkeypox virus E5, ... | | Authors: | Zhang, W, Liu, Y, Gao, H, Gan, J. | | Deposit date: | 2023-12-20 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional insights into the helicase protein E5 of Mpox virus.
Cell Discov, 10, 2024
|
|
3V1Z
 
 | | Crystal structure of Type IIF restriction endonuclease Bse634I with cognate DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*CP*G)-3'), Endonuclease Bse634IR | | Authors: | Manakova, E.N, Grazulis, S, Golovenko, D, Tamulaitiene, G. | | Deposit date: | 2011-12-11 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease.
Nucleic Acids Res., 40, 2012
|
|
6O1D
 
 | |
5ZOE
 
 | | Crystal Structure of D181A hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
8C7A
 
 | |
