6FL5
 
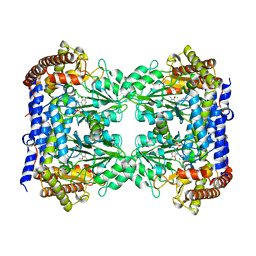 | | Structure of human SHMT1-H135N-R137A-E168N mutant at 3.6 Ang. resolution | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase, ... | | Authors: | Giardina, G, Cutruzzola, F, Lucchi, R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The catalytic activity of serine hydroxymethyltransferase is essential for de novo nuclear dTMP synthesis in lung cancer cells.
FEBS J., 285, 2018
|
|
6FJW
 
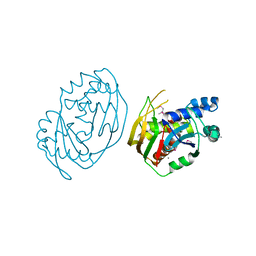 | |
6FN9
 
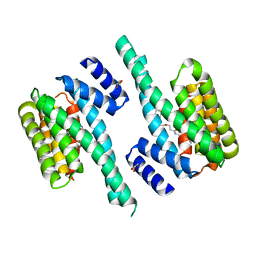 | |
6G73
 
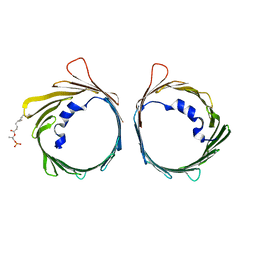 | | The dynamic nature of the VDAC1 channels in bilayers: human VDAC1 at 3.3 Angstrom resolution | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Razeto, A, Gribbon, P, Loew, C. | | Deposit date: | 2018-04-04 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | The dynamic nature of the VDAC1 channels in bilayers as revealed by two crystal structures of the human isoform in bicelles at 2.7 and 3.3 Angstrom resolution: implications for VDAC1 voltage-dependent mechanism and for its oligomerization
To Be Published
|
|
6G7O
 
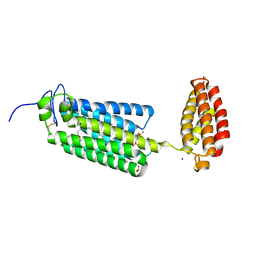 | | Crystal structure of human alkaline ceramidase 3 (ACER3) at 2.7 Angstrom resolution | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Alkaline ceramidase 3,Soluble cytochrome b562, CALCIUM ION, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Healey, R.D, Sounier, R, Grison, C, Hoh, F, Basu, S, Granier, S. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a human intramembrane ceramidase explains enzymatic dysfunction found in leukodystrophy.
Nat Commun, 9, 2018
|
|
6G8C
 
 | |
6FNB
 
 | |
6FPY
 
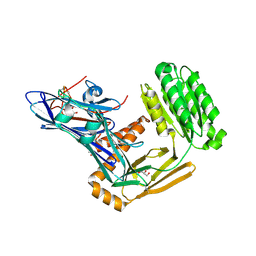 | | Inter-alpha-inhibitor heavy chain 1, wild type | | Descriptor: | GLYCEROL, Inter-alpha-trypsin inhibitor heavy chain H1, MAGNESIUM ION | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Inter-alpha-inhibitor heavy chain-1 has an integrin-like 3D structure mediating immune regulatory activities and matrix stabilization during ovulation
J.Biol.Chem., 2020
|
|
6FWK
 
 | |
6FXA
 
 | |
3JXC
 
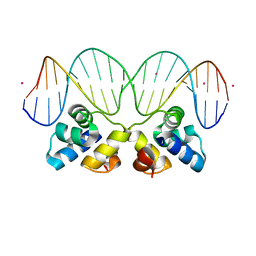 | |
3K1Y
 
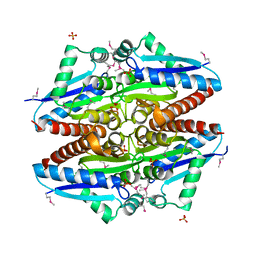 | | X-ray structure of oxidoreductase from corynebacterium diphtheriae. orthorombic crystal form, northeast structural genomics consortium target cdr100d | | Descriptor: | SULFATE ION, oxidoreductase | | Authors: | Kuzin, A, Lew, S, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-20 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target CdR100D
To be Published
|
|
3K3W
 
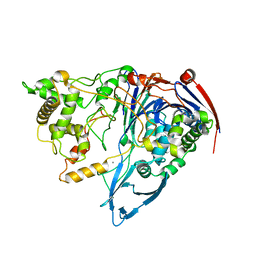 | | Thermostable Penicillin G acylase from Alcaligenes faecalis in orthorhombic form | | Descriptor: | CALCIUM ION, Penicillin G acylase | | Authors: | Varshney, N.K, Kumar, R.S, Ignatova, Z, Dodson, E, Suresh, C.G. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystallization and X-ray structure analysis of a thermostable penicillin G acylase from Alcaligenes faecalis.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3JAR
 
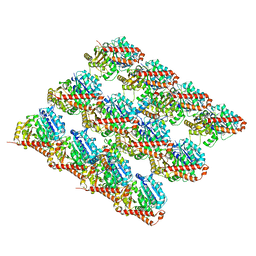 | | Cryo-EM structure of GDP-microtubule co-polymerized with EB3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, R, Nogales, E. | | Deposit date: | 2015-06-19 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins.
Cell(Cambridge,Mass.), 162, 2015
|
|
3K03
 
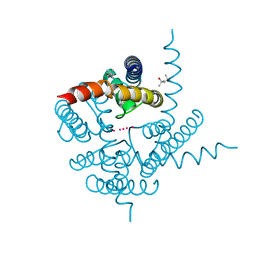 | | Crystal Structure of CNG mimicking NaK mutant, NaK-DTPP, K+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein NaK | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K3P
 
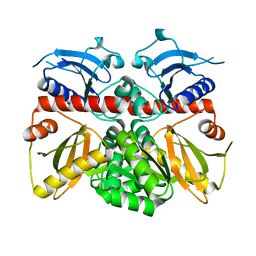 | |
3JPY
 
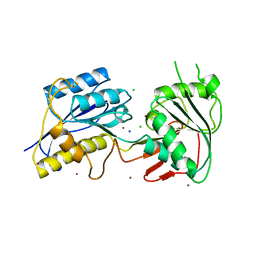 | | Crystal structure of the zinc-bound amino terminal domain of the NMDA receptor subunit NR2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutamate [NMDA] receptor subunit epsilon-2, ... | | Authors: | Karakas, E, Simorowski, N, Furukawa, H. | | Deposit date: | 2009-09-04 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit.
Embo J., 28, 2009
|
|
3K6I
 
 | |
3JRN
 
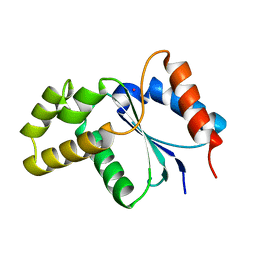 | | Crystal structure of TIR domain from Arabidopsis Thaliana | | Descriptor: | ARSENIC, AT1G72930 protein | | Authors: | Chan, S.L, Mukasa, T, Santelli, E, Low, L.Y, Pascual, J. | | Deposit date: | 2009-09-08 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants.
Protein Sci., 19, 2009
|
|
3JTC
 
 | | Importance of Mg2+ in the Ca2+-Dependent Folding of the gamma-Carboxyglutamic Acid Domains of Vitamin K-Dependent clotting and anticlotting Proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endothelial protein C receptor, ... | | Authors: | Bajaj, S.P, Vadivel, K, Agah, S, Cascio, D, Krishnaswamy, S, Esmon, C, Padmanabhan, K. | | Deposit date: | 2009-09-11 | | Release date: | 2011-04-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Studies of gamma-Carboxyglutamic Acid Domains of Factor VIIa and Activated Protein C: Role of Magnesium at Physiological Calcium.
J.Mol.Biol., 425, 2013
|
|
3JXB
 
 | |
3K3R
 
 | |
3K5R
 
 | |
3K6D
 
 | |
3K87
 
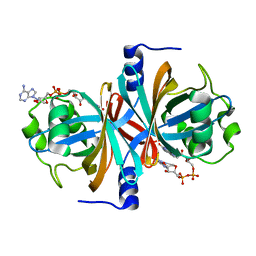 | | Crystal structure of NADH:FAD oxidoreductase (TftC) - FAD complex | | Descriptor: | Chlorophenol-4-monooxygenase component 1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kang, C.H, Webb, B.N. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of chlorophenol 4-monooxygenase (TftD) and NADH:FAD oxidoreductase (TftC) of Burkholderia cepacia AC1100.
J.Biol.Chem., 285, 2010
|
|
