6WE2
 
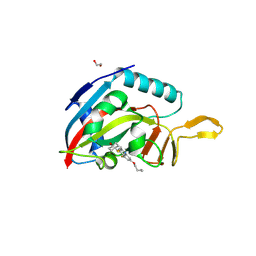 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN012759 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylmethoxy)-5-fluoro-2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}quinazolin-4(3H)-one, Isoform 1 of Protein mono-ADP-ribosyltransferase PARP14 | | Authors: | Swinger, K.K, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
6WZS
 
 | | Fusibacterium ulcerans ZTP riboswitch bound to m-1-pyridinyl AICA | | Descriptor: | 5-amino-1-(pyridin-3-yl)-1H-imidazole-4-carboxamide, Fusibacterium ulcerans ZTP riboswitch, MAGNESIUM ION, ... | | Authors: | Pichling, P, Jones, C.P, Ferre-D'Amare, A.R, Tran, B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Parallel Discovery Strategies Provide a Basis for Riboswitch Ligand Design.
Cell Chem Biol, 27, 2020
|
|
6WSM
 
 | | Crystal structure of coiled coil region of human septin 8 | | Descriptor: | SULFATE ION, Septin-8 | | Authors: | Cabrejos, D.A.L, Cavini, I, Sala, F.A, Valadares, N.F, Pereira, H.M, Brandao-Neto, J, Nascimento, A.F.Z, Uson, I, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Orientational Ambiguity in Septin Coiled Coils and its Structural Basis.
J.Mol.Biol., 433, 2021
|
|
6WZR
 
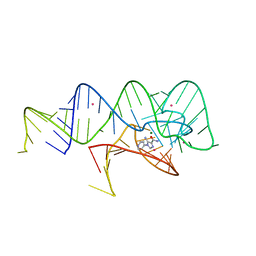 | | Fusibacterium ulcerans ZTP riboswitch bound to p-1-pyridinyl AICA | | Descriptor: | 5-amino-1-(pyridin-4-yl)-1H-imidazole-4-carboxamide, Fusibacterium ulcerans ZTP riboswitch, MAGNESIUM ION, ... | | Authors: | Pichling, P, Jones, C.P, Ferre-D'Amare, A.R, Tran, B. | | Deposit date: | 2020-05-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Parallel Discovery Strategies Provide a Basis for Riboswitch Ligand Design.
Cell Chem Biol, 27, 2020
|
|
2YTI
 
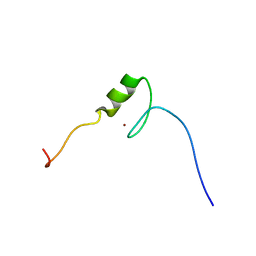 | | Solution structure of the C2H2 type zinc finger (region 564-596) of human Zinc finger protein 347 | | Descriptor: | ZINC ION, Zinc finger protein 347 | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 564-596) of human Zinc finger protein 347
To be Published
|
|
2YTU
 
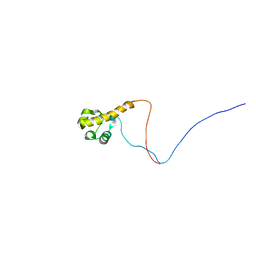 | | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor | | Descriptor: | Friend leukemia integration 1 transcription factor | | Authors: | Goroncy, A.K, Sato, M, Tochio, N, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor
To be Published
|
|
2YUK
 
 | | Solution structure of the HMG box of human Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog | | Descriptor: | Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog | | Authors: | Abe, H, Tochio, N, Miyamoto, K, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG box of human Myeloid/lymphoid or mixed-lineage leukemia protein 3 homolog
To be Published
|
|
2YUV
 
 | | Solution Structure of 2nd Immunoglobulin Domain of Slow Type Myosin-Binding Protein C | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Niraula, T.N, Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 2nd Immunoglobulin Domain of Slow Type Myosin-Binding Protein C
To be Published
|
|
6WE4
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one, 8-methyl-2-{[(pyridin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, ... | | Authors: | Swinger, K.S, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
7C6B
 
 | | Crystal structure of Ago2 MID domain in complex with 6-(3-(2-carboxyethyl)phenyl)purine riboside monophosphate | | Descriptor: | 3-[3-[9-[(2R,3R,4S,5R)-3,4-bis(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]purin-6-yl]phenyl]propanoic acid, PHOSPHATE ION, Protein argonaute-2 | | Authors: | Suzuki, M, Takahashi, Y, Saito, J, Miyagi, H, Shinohara, F. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | siRNA potency enhancement via chemical modifications of nucleotide bases at the 5'-end of the siRNA guide strand.
Rna, 27, 2021
|
|
6WSI
 
 | | Intact cis-2,3-epoxysuccinic acid bound to Isocitrate Lyase-1 from Mycobacterium tuberculosis | | Descriptor: | (2R,3S)-oxirane-2,3-dicarboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Krieger, I.V, Mellott, D, Meek, T.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Covalent Inactivation of Mycobacterium tuberculosis Isocitrate Lyase by cis -2,3-Epoxy-Succinic Acid.
Acs Chem.Biol., 16, 2021
|
|
6DP7
 
 | |
2YWG
 
 | | Crystal structure of GTP-bound LepA from Aquifex aeolicus | | Descriptor: | GTP-binding protein LepA, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Nishino, A, Nakayama-Ushikoshi, R, Hanawa-Suetsugu, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structures of GTP-binding protein LepA from Aquifex aeolicus
To be Published
|
|
4K3A
 
 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | SULFATE ION, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6WCG
 
 | |
6WCN
 
 | |
6WCP
 
 | |
2YXG
 
 | |
2YYE
 
 | | Crystal structure of selenophosphate synthetase from Aquifex aeolicus complexed with AMPCPP | | Descriptor: | COBALT (II) ION, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, PHOSPHATE ION, ... | | Authors: | Itoh, Y, Sekine, S, Matsumoto, E, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-29 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of selenophosphate synthetase essential for selenium incorporation into proteins and RNAs
J.Mol.Biol., 385, 2009
|
|
2YYW
 
 | |
6WCL
 
 | |
6WI7
 
 | | RING1B-BMI1 fusion in closed conformation | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase RING2, Polycomb complex protein BMI-1 chimera, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Small-molecule inhibitors targeting Polycomb repressive complex 1 RING domain.
Nat.Chem.Biol., 17, 2021
|
|
6WCM
 
 | |
2YZ8
 
 | | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556) | | Descriptor: | Obscurin | | Authors: | Saijo, S, Ohsawa, N, Nishino, A, Kishishita, S, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the 32th Ig-like domain of human obscurin (KIAA1556)
To be Published
|
|
6WCH
 
 | |
