3BSD
 
 | |
3BYV
 
 | | Crystal structure of Toxoplasma gondii specific rhoptry antigen kinase domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Rhoptry kinase | | Authors: | Wernimont, A.K, Lunin, V.V, Yang, C, Lew, J, Kozieradzki, I, Lin, Y.H, Sun, X, Khuu, C, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bochkarev, A, Hui, R, Sibley, D, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-16 | | Release date: | 2008-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii.
Embo J., 28, 2009
|
|
3CBF
 
 | | Crystal structure of LysN, alpha-aminoadipate aminotransferase, from Thermus thermophilus HB27 | | Descriptor: | (2S)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]hexanedioic acid, Alpha-aminodipate aminotransferase | | Authors: | Tomita, T, Miyazaki, T, Miyagawa, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2008-02-21 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mechanism for multiple-substrates recognition of alpha-aminoadipate aminotransferase from Thermus thermophilus
Proteins, 2008
|
|
3BZE
 
 | | The human non-classical major histocompatibility complex molecule HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Hoare, H.L, Sullivan, L.C, Ely, L.K, Beddoe, T, Henderson, K.N, Lin, J, Clements, C.S, Reid, H.H, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2008-01-17 | | Release date: | 2008-04-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors
J.Mol.Biol., 377, 2008
|
|
2UUY
 
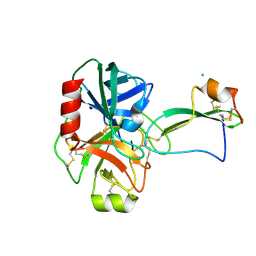 | | Structure of a tick tryptase inhibitor in complex with bovine trypsin | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, CHLORIDE ION, ... | | Authors: | Siebold, C, Paesen, G.C, Harlos, K, Peacey, M.F, Nuttall, P.A, Stuart, D.I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Tick Protein with a Modified Kunitz Fold Inhibits Human Tryptase.
J.Mol.Biol., 368, 2007
|
|
1YHM
 
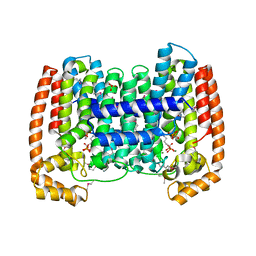 | | Structure of the complex of Trypanosoma cruzi farnesyl disphosphate synthase with alendronate, Isopentenyl diphosphate and mg+2 | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, MAGNESIUM ION, ... | | Authors: | Gabelli, S.B, McLellan, J.S, Montalvetti, A, Oldfield, E, Docampo, R, Amzel, L.M. | | Deposit date: | 2005-01-09 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the farnesyl diphosphate synthase from Trypanosoma cruzi: Implications for drug design.
Proteins, 62, 2005
|
|
3S32
 
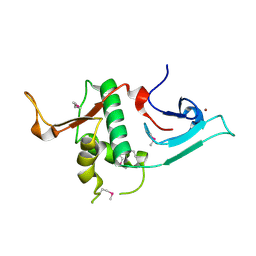 | | Crystal structure of Ash2L N-terminal domain | | Descriptor: | Set1/Ash2 histone methyltransferase complex subunit ASH2, ZINC ION | | Authors: | Sarvan, S, Avdic, V, Tremblay, V, Chaturvedi, C.-P, Zhang, P, Lanouette, S, Blais, A, Brunzelle, J.S, Brand, M, Couture, J.-F. | | Deposit date: | 2011-05-17 | | Release date: | 2011-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the trithorax group protein ASH2L reveals a forkhead-like DNA binding domain.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4IAH
 
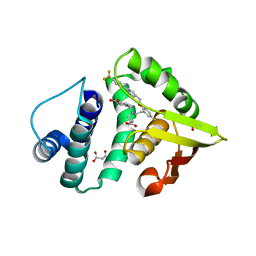 | | Crystal Structure of BAY 60-2770 bound C139A H-NOX domain with S-nitrosylated conserved C122 | | Descriptor: | 4-({(4-carboxybutyl)[2-(5-fluoro-2-{[4'-(trifluoromethyl)biphenyl-4-yl]methoxy}phenyl)ethyl]amino}methyl)benzoic acid, Alr2278 protein, MALONATE ION | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2012-12-06 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into BAY 60-2770 Activation and S-Nitrosylation-Dependent Desensitization of Soluble Guanylyl Cyclase via Crystal Structures of Homologous Nostoc H-NOX Domain Complexes.
Biochemistry, 52, 2013
|
|
3I6Y
 
 | | Structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
2OG0
 
 | | Crystal Structure of the Lambda Xis-DNA complex | | Descriptor: | 5'-D(*AP*AP*AP*CP*AP*GP*AP*CP*TP*AP*CP*AP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*TP*AP*TP*TP*AP*TP*GP*TP*AP*GP*TP*CP*TP*GP*TP*TP*T)-3', Excisionase | | Authors: | Papagiannis, C.V, Sam, M.D, Abbani, M.A, Cascio, D, Yoo, D, Clubb, R.T, Johnson, R.C. | | Deposit date: | 2007-01-04 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fis targets assembly of the xis nucleoprotein filament to promote excisive recombination by phage lambda.
J.Mol.Biol., 367, 2007
|
|
2OYY
 
 | | HTHP: a hexameric tyrosine-coordinated heme protein | | Descriptor: | Hexameric cytochrome, IODIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dobbek, H. | | Deposit date: | 2007-02-23 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | HTHP: A Novel Class of Hexameric, Tyrosine-coordinated Heme Proteins
J.Mol.Biol., 368, 2007
|
|
4X63
 
 | | Crystal structure of PRMT5:MEP50 with EPZ015666 and SAH | | Descriptor: | Methylosome protein 50, N-[(2S)-3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl]-6-(oxetan-3-ylamino)pyrimidine-4-carboxamide, Protein arginine N-methyltransferase 5, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Nat.Chem.Biol., 11, 2015
|
|
6CZN
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Z2OHTPE and a glucocorticoid receptor-interacting protein 1 NR box II peptide | | Descriptor: | 4,4'-[(1R,2R)-1-phenylbutane-1,2-diyl]diphenol, Estrogen receptor, GRIP Peptide | | Authors: | Fanning, S.W, Han, R, Maximov, P, Jordan, V.C, Greene, G.L. | | Deposit date: | 2018-04-09 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Z2OHTPE and a glucocorticoid receptor-interacting protein 1 NR box II peptide
To Be Published
|
|
4XG3
 
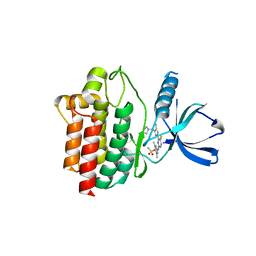 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 4-{[5-fluoro-4-(3-{[(3R)-3-hydroxypyrrolidin-1-yl]methyl}-4-methyl-1H-pyrrol-1-yl)pyrimidin-2-yl]amino}-2,6-dimethylphenyl methanesulfonate, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG7
 
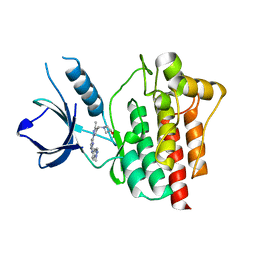 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-[(3-methyl-1-{2-[(1-methyl-1H-indazol-5-yl)amino]pyrimidin-4-yl}-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG6
 
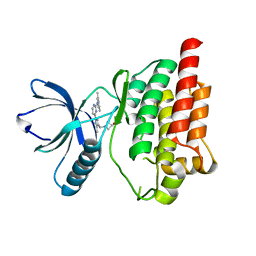 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-[(1-{2-[(3,5-dimethylphenyl)amino]pyrimidin-4-yl}-3-methyl-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
6ITK
 
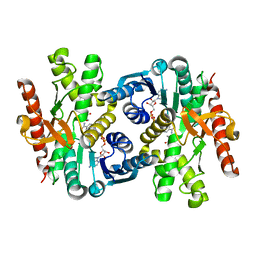 | |
4XG4
 
 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | (3R)-1-{[1-(5-fluoro-2-{[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]amino}pyrimidin-4-yl)-4-methyl-1H-pyrrol-3-yl]methyl}pyrrolidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
4XG9
 
 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-[(1-{2-[(3-chloro-1,2-dimethyl-1H-indol-5-yl)amino]pyrimidin-4-yl}-3-methyl-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
5A3N
 
 | | Crystal structure of human PLU-1 (JARID1B) in complex with KDOAM25a | | Descriptor: | 1,2-ETHANEDIOL, 2-[[[2-[2-(dimethylamino)ethyl-ethyl-amino]-2-oxidanylidene-ethyl]amino]methyl]pyridine-4-carboxamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Nuzzi, A, Ruda, G.F, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Brennan, P, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective KDM5 Inhibitor Stops Cellular Demethylation of H3K4me3 at Transcription Start Sites and Proliferation of MM1S Myeloma Cells.
Cell Chem Biol, 24, 2017
|
|
5BS0
 
 | | MAGE-A3 Reactive TCR in complex with Titin Epitope in HLA-A1 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Raman, M.C.C, Rizkallah, P.J, Simmons, R, Donellan, Z, Dukes, J, Bossi, G, LeProvost, G, Mahon, T, Hickman, E, Lomax, M, Oates, J, Hassan, N, Vuidepot, A, Sami, M, Cole, D.K, Jakobsen, B.K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Direct molecular mimicry enables off-target cardiovascular toxicity by an enhanced affinity TCR designed for cancer immunotherapy.
Sci Rep, 6, 2016
|
|
5BRZ
 
 | | MAGE-A3 reactive TCR in complex with MAGE-A3 in HLA-A1 | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-PRO-ILE-GLY-HIS-LEU-TYR, HLA class I histocompatibility antigen, ... | | Authors: | Raman, M.C.C, Rizkallah, P.J, Simmons, R, Donnellan, Z, Dukes, J, Bossi, G, LeProvost, G, Mahon, T, Hickman, E, LomaX, M, Oates, J, Hassan, N, Vuidepot, A, Sami, M, Cole, D.K, Jakobsen, B.K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Direct molecular mimicry enables off-target cardiovascular toxicity by an enhanced affinity TCR designed for cancer immunotherapy.
Sci Rep, 6, 2016
|
|
5B1S
 
 | | Crystal structure of Trypanosoma cruzi spermidine synthase in complex with 2-(2-fluorophenyl)ethanamine | | Descriptor: | 2-(2-fluorophenyl)ethanamine, 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, Spermidine synthase, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | In silico, in vitro, X-ray crystallography, and integrated strategies for discovering spermidine synthase inhibitors for Chagas disease
Sci Rep, 7, 2017
|
|
4X60
 
 | | Crystal structure of PRMT5:MEP50 with EPZ015666 and sinefungin | | Descriptor: | GLYCEROL, Methylosome protein 50, N-[(2S)-3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl]-6-(oxetan-3-ylamino)pyrimidine-4-carboxamide, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-06 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Nat.Chem.Biol., 11, 2015
|
|
6OFK
 
 | | Crystal structure of green fluorescent protein (GFP); S65T; ih circular permutant (50-51) | | Descriptor: | ACETATE ION, Green Fluorescent Protein (GFP); S65T; ih circular permutant (50-51) | | Authors: | Lin, C.-Y, Romei, M.G, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2019-03-30 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Unified Model for Photophysical and Electro-Optical Properties of Green Fluorescent Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
