4E77
 
 | | 2.0A Crystal Structure of a Glutamate-1-Semialdehyde Aminotransferase from Yersinia pestis CO92 | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase, NITRATE ION, SODIUM ION | | Authors: | Brunzelle, J.S, Wawrzak, W, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-16 | | Release date: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A Crystal Structure of a Glutamate-1-Semialdehyde Aminotransferase from Yersinia pestis CO92
TO BE PUBLISHED
|
|
4ECO
 
 | |
2H6V
 
 | |
4ECN
 
 | |
4EGS
 
 | |
4CQH
 
 | | Structure of Infrared Fluorescent Protein IFP2.0 | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, BACTERIOPHYTOCHROME, SODIUM ION | | Authors: | Lafaye, C, Yu, D, Noirclerc-Savoye, M, Shu, X, Royant, A. | | Deposit date: | 2014-02-17 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | An Improved Monomeric Infrared Fluorescent Protein for Neuronal and Tumour Brain Imaging.
Nat.Commun., 5, 2014
|
|
4CFY
 
 | | SAVINASE CRYSTAL STRUCTURES FOR COMBINED SINGLE CRYSTAL DIFFRACTION AND POWDER DIFFRACTION ANALYSIS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN SAVINASE | | Authors: | Frankaer, C.G, Moroz, O.V, Turkenburg, J.P, Aspmo, S.I, Thymark, M, Friis, E.P, Stahla, K, Nielsen, J.E, Wilson, K.S, Harris, P. | | Deposit date: | 2013-11-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Analysis of an Industrial Production Suspension of Bacillus Lentus Subtilisin Crystals by Powder Diffraction: A Powerful Quality-Control Tool.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CBX
 
 | | Crystal structure of Plasmodium berghei actin II | | Descriptor: | ACTIN-2, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Vahokoski, J, Bhargav, S.P, Desfosses, A, Andreadaki, M, Kumpula, E.P, Ignatev, A, Munico Martinez, S, Lepper, S, Frischknecht, F, Siden-Kiamos, I, Sachse, C, Kursula, I. | | Deposit date: | 2013-10-17 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Differences Explain Diverse Functions of Plasmodium Actins.
Plos Pathog., 10, 2014
|
|
4CCG
 
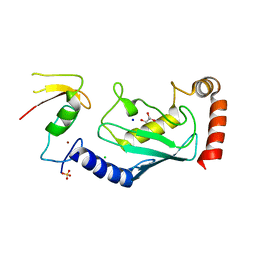 | | Structure of an E2-E3 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, CHLORIDE ION, ... | | Authors: | Hodson, C, Purkiss, A, Walden, H. | | Deposit date: | 2013-10-22 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Human Fancl Ring-Ube2T Complex Reveals Determinants of Cognate E3-E2 Selection.
Structure, 22, 2014
|
|
2GL3
 
 | |
4EUH
 
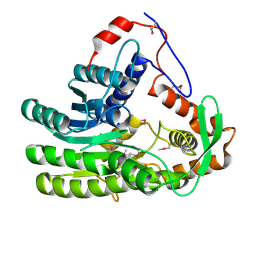 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase apo form | | Descriptor: | Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
4EW7
 
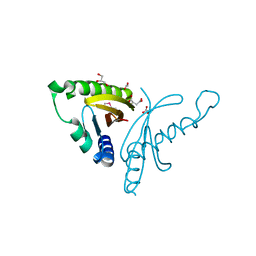 | | The crystal structure of conjugative transfer PAS_like domain from Salmonella enterica subsp. enterica serovar Typhimurium | | Descriptor: | ACETIC ACID, CHLORIDE ION, Conjugative transfer: regulation, ... | | Authors: | Wu, R, Jedrzejczak, R.P, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-04-26 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The crystal structure of conjugative transfer PAS_like domain from Salmonella enterica subsp. enterica serovar Typhimurium
To be Published
|
|
4FE3
 
 | |
4F06
 
 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 RPB_2270 in complex with P-HYDROXYBENZOIC ACID | | Descriptor: | Extracellular ligand-binding receptor, GLYCEROL, P-HYDROXYBENZOIC ACID, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-15 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
2HTI
 
 | |
4F6U
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 5 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[3-(morpholin-4-yl)propyl]urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-[3-(morpholin-4-yl)propyl]urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2HRW
 
 | | Crystal Structure of Phosphonopyruvate Hydrolase | | Descriptor: | CHLORIDE ION, Phosphonopyruvate hydrolase, SODIUM ION | | Authors: | Chen, C.C.H, Herzberg, O. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Kinetics of Phosphonopyruvate Hydrolase from Voriovorax sp. Pal2: New Insight into the Divergence of Catalysis within the PEP Mutase/Isocitrate Lyase Superfamily
Biochemistry, 45, 2006
|
|
2JGP
 
 | | Structure of the TycC5-6 PCP-C bidomain of the tyrocidine synthetase TycC | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, SULFATE ION, ... | | Authors: | Samel, S.A, Schoenafinger, G, Knappe, T.A, Marahiel, M.A, Essen, L.-O. | | Deposit date: | 2007-02-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Functional Insights Into a Peptide Bond-Forming Bidomain from a Nonribosomal Peptide Synthetase.
Structure, 15, 2007
|
|
2JH6
 
 | | Human Thrombin Hirugen Inhibitor complex | | Descriptor: | 2-(5-CHLORO-2-THIENYL)-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}ETHANESULFONAMIDE, CALCIUM ION, HIRUDIN IIIA, ... | | Authors: | Senger, S, Chan, C, Convery, M.A, Hubbard, J.A, Young, R.J, Shah, G.P, Watson, N.S. | | Deposit date: | 2007-02-20 | | Release date: | 2007-05-08 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Sulfonamide-Related Conformational Effects and Their Importance in Structure-Based Design.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3UU5
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-20' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UWP
 
 | | Crystal structure of Dot1l in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-02 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
3UR7
 
 | | Higher-density crystal structure of potato endo-1,3-beta-glucanase | | Descriptor: | Glucan endo-1,3-beta-D-glucosidase, SODIUM ION | | Authors: | Wojtkowiak, A, Witek, K, Hennig, J, Jaskolski, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two high-resolution structures of potato endo-1,3-beta-glucanase reveal subdomain flexibility with implications for substrate binding
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UQ2
 
 | | Crystal structure of the post-catalytic product complex of polymerase lambda with an rCMP inserted opposite a templating G and dAMP inserted opposite a templating T at the primer terminus. | | Descriptor: | 5'-D(*CP*AP*GP*TP*AP)-R(P*CP*A)-3', 5'-D(*CP*GP*GP*CP*TP*GP*TP*AP*CP*TP*G)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Gosavi, R.A, Moon, A.F, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The catalytic cycle for ribonucleotide incorporation by human DNA Pol lambda
Nucleic Acids Res., 40, 2012
|
|
3UU9
 
 | | Structure of the free TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase | | Descriptor: | CALCIUM ION, Eight-heme nitrite reductase, HEME C, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2YME
 
 | | Crystal structure of a mutant binding protein (5HTBP-AChBP) in complex with granisetron | | Descriptor: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Kesters, D, Thompson, A.J, Brams, M, Elk, R.v, Spurny, R, Geitmann, M, Villalgordo, J.M, Guskov, A, Danielson, U.H, Lummis, S.C.R, Smit, A.B, Ulens, C. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Ligand Recognition in 5-Ht3 Receptors.
Embo Rep., 14, 2013
|
|
