6VOP
 
 | | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Escherichia coli | | Descriptor: | Aldolase | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-31 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Escherichia coli
To Be Published
|
|
6VOQ
 
 | | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Klebsiella pneumoniae | | Descriptor: | Aldolase, CHLORIDE ION, ZINC ION | | Authors: | Stogios, P.J, Evdokimova, E, McChesney, C, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-31 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of YgbL, a putative aldolase/epimerase/decarboxylase from Klebsiella pneumoniae
To Be Published
|
|
6OFO
 
 | | Crystal structure of split green fluorescent protein (GFP); s10 circular permutant (194-195) | | Descriptor: | Green fluorescent protein (GFP); s10 circular permutant (194-195) | | Authors: | Lin, C.-Y, Romei, M.G, Deller, M.C, Doukov, T.I, Boxer, S.G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Unified Model for Photophysical and Electro-Optical Properties of Green Fluorescent Proteins.
J.Am.Chem.Soc., 141, 2019
|
|
6VPG
 
 | |
6OXX
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-18 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6VSX
 
 | |
6PXQ
 
 | | Crystal structure of human thrombin mutant D194A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin heavy chain, Thrombin light chain | | Authors: | Stojanovski, B, Chen, Z, Koester, S.K, Pelc, L.A, Di Cera, E. | | Deposit date: | 2019-07-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Role of the I16-D194 ionic interaction in the trypsin fold.
Sci Rep, 9, 2019
|
|
6PTA
 
 | | Crystal structure of the ARF family small GTPase ARF1 from Candida albicans in complex with GDP | | Descriptor: | ADP-ribosylation factor, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-15 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the ARF family small GTPase ARF1 from Candida albicans in complex with GDP
To Be Published
|
|
2WQX
 
 | | InlB321_4R: S199R, D200R, G206R, A227R, C242A mutant of the Listeria monocytogenes InlB internalin domain | | Descriptor: | INTERNALIN B | | Authors: | Niemann, H.H, Ferraris, D.M, Heinz, D.W. | | Deposit date: | 2009-08-27 | | Release date: | 2009-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand-Mediated Dimerization of the met Receptor Tyrosine Kinase by the Bacterial Invasion Protein Inlb.
J.Mol.Biol., 395, 2010
|
|
6Q2K
 
 | |
1YH2
 
 | | Ubiquitin-Conjugating Enzyme HSPC150 | | Descriptor: | HSPC150 protein similar to ubiquitin-conjugating enzyme | | Authors: | Walker, J.R, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
6Q1P
 
 | | Glucocerebrosidase in complex with pharmacological chaperone norIMX8 | | Descriptor: | (2S,3S,4S,5R)-2-{[(4-methylpentyl)sulfonyl]methyl}piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vickers, C, Withers, S.G, Boraston, A.B. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Pharmacological chaperones for GCase that switch conformation with pH enhance enzyme levels in Gaucher animal models.
To Be Published
|
|
6VPB
 
 | |
6OPU
 
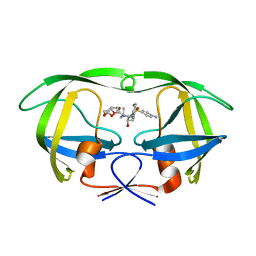 | | HIV-1 Protease NL4-3 K45I, M46I, V82F, I84V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease NL4-3 | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2019-04-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Picomolar to Micromolar: Elucidating the Role of Distal Mutations in HIV-1 Protease in Conferring Drug Resistance.
Acs Chem.Biol., 14, 2019
|
|
6OIE
 
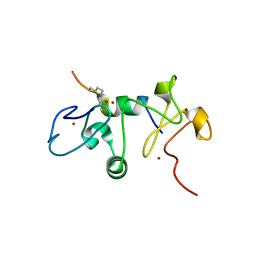 | |
6OXQ
 
 | | HIV-1 Protease NL4-3 WT in Complex with UMass8 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-3-[(2-ethylbutyl){[4-(hydroxymethyl)phenyl]sulfonyl}amino]-2-hydroxypropyl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXV
 
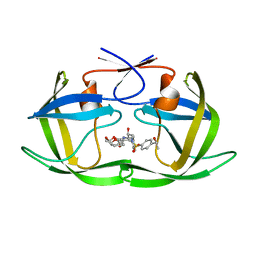 | | HIV-1 Protease NL4-3 WT in Complex with LR-85 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[(2-ethylbutyl)({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OYG
 
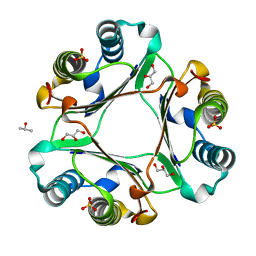 | |
6OVJ
 
 | |
6OXY
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-19 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OY8
 
 | |
6OL4
 
 | | Protein Tyrosine Phosphatase 1B (1-301), F182A mutant, apo state | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Cui, D.S, Lipchock, J.M, Loria, J.P. | | Deposit date: | 2019-04-15 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
6OOU
 
 | | Crystal structure of HIV-1 Protease NL4-3 L89V Mutant in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, NL4-3 PROTEASE, SULFATE ION | | Authors: | Henes, M, Kosovrasti, K, Lockbaum, G.J, Leidner, F, Nachum, G.S, Nalivaika, E.A, Bolon, D.N.A, KurtYilmaz, N, Schiffer, C.A, Whitfield, T.W. | | Deposit date: | 2019-04-23 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Molecular Determinants of Epistasis in HIV-1 Protease: Elucidating the Interdependence of L89V and L90M Mutations in Resistance.
Biochemistry, 58, 2019
|
|
6O56
 
 | | HNH Nuclease from S. pyogenes Cas9 | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1 | | Authors: | Newton, J.C, Lisi, G.P, Jogl, G. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric Motions of the CRISPR-Cas9 HNH Nuclease Probed by NMR and Molecular Dynamics.
J.Am.Chem.Soc., 142, 2020
|
|
6O62
 
 | | Crystal structure of Sec4p, a Rab family GTPase from Candida albicans | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Ras-related protein SEC4 | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-03-05 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | To be published
To Be Published
|
|
