4UCF
 
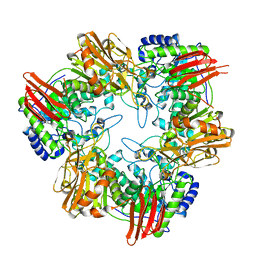 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, N-PROPANOL, ... | | Authors: | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpha-Galactose.
FEBS J., 283, 2016
|
|
4UE3
 
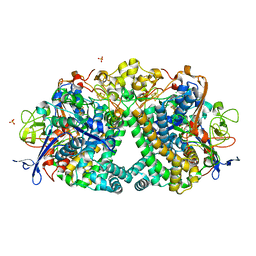 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases and the Importance of the active site Arginine | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
4UDY
 
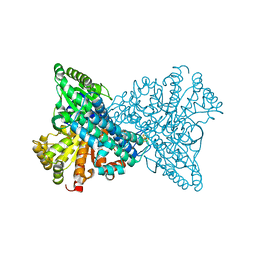 | | NCO- bound to cluster C of Ni,Fe-CO dehydrogenase at true-atomic resolution | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE 2, FE (II) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Fesseler, J, Jeoung, J.-H, Dobbek, H. | | Deposit date: | 2014-12-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | How the [Nife4 S4 ] Cluster of Co Dehydrogenase Activates Co2 and Nco(.)
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4U40
 
 | |
4U42
 
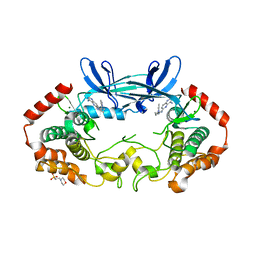 | | MAP4K4 T181E Mutant Bound to inhibitor compound 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[(3S)-3-(4-methyl-1H-pyrazol-3-yl)piperidin-1-yl]pyrido[3,2-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U46
 
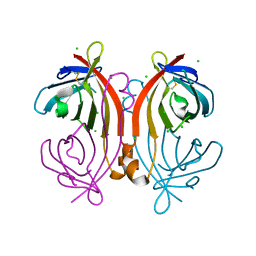 | | Crystal structure of an avidin mutant | | Descriptor: | Avidin, CHLORIDE ION | | Authors: | Agrawal, N, Lehtonen, S, Kahkonen, N, Riihimaki, T, Hytonen, V.P, Kulomaa, M.S, Johnson, M.S, Airenne, T.T. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Artificial Avidin-Based Receptors for a Panel of Small Molecules.
Acs Chem.Biol., 11, 2016
|
|
4U2N
 
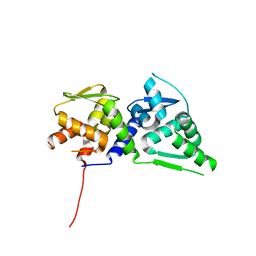 | |
4U2Q
 
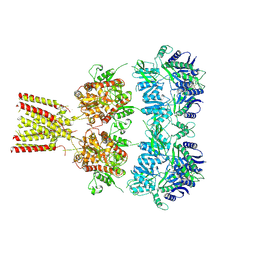 | | Full-length AMPA subtype ionotropic glutamate receptor GluA2 in complex with partial agonist kainate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Duerr, K.L, Chen, L, Gouaux, E. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5247 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
4U3D
 
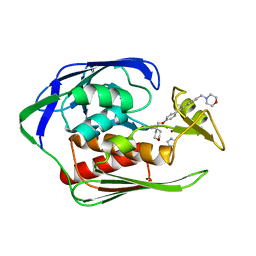 | |
4U4B
 
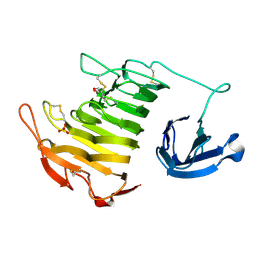 | |
4U4C
 
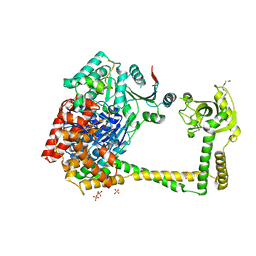 | | The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities | | Descriptor: | 1,2-ETHANEDIOL, ATP-dependent RNA helicase DOB1, CHLORIDE ION, ... | | Authors: | Falk, S, Weir, J.R, Hentschel, J, Reichelt, P, Bonneau, F, Conti, E. | | Deposit date: | 2014-07-23 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Molecular Architecture of the TRAMP Complex Reveals the Organization and Interplay of Its Two Catalytic Activities.
Mol.Cell, 55, 2014
|
|
4U4I
 
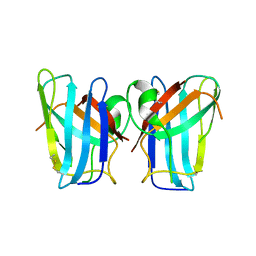 | | Megavirus chilensis superoxide dismutase | | Descriptor: | Cu/Zn superoxide dismutase | | Authors: | Lartigue, A, Claverie, J.-M, Burlat, B, Coutard, B, Abergel, C. | | Deposit date: | 2014-07-23 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The megavirus chilensis cu,zn-superoxide dismutase: the first viral structure of a typical cellular copper chaperone-independent hyperstable dimeric enzyme.
J.Virol., 89, 2015
|
|
4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
4U3H
 
 | | Crystal structure of FN3con | | Descriptor: | FN3con | | Authors: | Porebski, B.T, McGowan, S, Buckle, A.M. | | Deposit date: | 2014-07-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and dynamic properties that govern the stability of an engineered fibronectin type III domain.
Protein Eng.Des.Sel., 28, 2015
|
|
4U3J
 
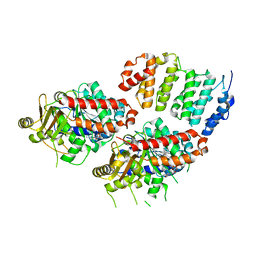 | | TOG2:alpha/beta-tubulin complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | Authors: | Ayaz, P, Rice, L.M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A tethered delivery mechanism explains the catalytic action of a microtubule polymerase.
Elife, 3, 2014
|
|
4U3T
 
 | |
4U3Z
 
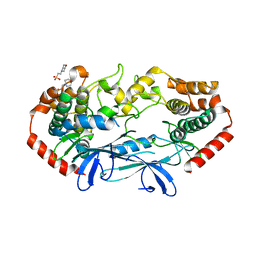 | | APO MAP4K4 T181E Phosphomimetic Mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mitogen-activated protein kinase kinase kinase kinase 4, SODIUM ION | | Authors: | Harris, S.F, Wu, P, Coons, M. | | Deposit date: | 2014-07-23 | | Release date: | 2016-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Plasticity and Kinase Activation in a Cohort of MAP4K4 Structures
to be published
|
|
4U47
 
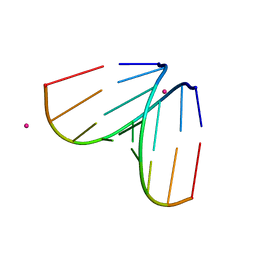 | | Octameric RNA duplex soaked in terbium(III)chloride | | Descriptor: | RNA (5'-R(*UP*CP*GP*UP*AP*CP*GP*A)-3'), TERBIUM(III) ION | | Authors: | Schaffer, M.F, Spingler, B, Schnabl, J, Peng, G, Olieric, V, Sigel, R.K.O. | | Deposit date: | 2014-07-23 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations.
Int J Mol Sci, 17, 2016
|
|
4U49
 
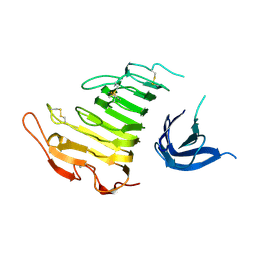 | |
4U4E
 
 | |
4U4H
 
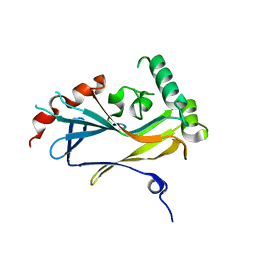 | |
4U4J
 
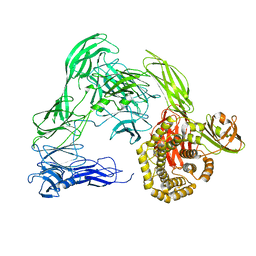 | |
4U4R
 
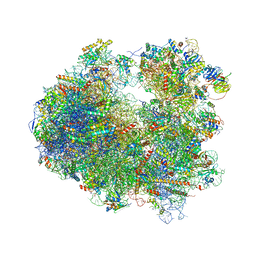 | | Crystal structure of Lactimidomycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R,5S,6E)-2-hydroxy-5-methyl-7-[(2R,3S,4E,6Z,10E)-3-methyl-12-oxooxacyclododeca-4,6,10-trien-2-yl]-4-oxooct-6-en-1-yl}piperidine-2,6-dione, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U5J
 
 | | C-Src in complex with Ruxolitinib | | Descriptor: | (3R)-3-cyclopentyl-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1H-pyrazol-1-yl]propanenitrile, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Chen, Y, Duan, Y, Chen, L. | | Deposit date: | 2014-07-25 | | Release date: | 2014-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | c-Src Binds to the Cancer Drug Ruxolitinib with an Active Conformation
Plos One, 9, 2014
|
|
4U5L
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzyl]-L-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
