1TUQ
 
 | | NMR Structure Analysis of the B-DNA Dodecamer CTCtCACGTGGAG with a tricyclic cytosin base analogue | | Descriptor: | 5'-D(P*CP*TP*CP*(TC1)P*AP*CP*GP*TP*GP*GP*AP*G)-3' | | Authors: | Engman, K.C, Sandin, P, Osborne, S, Brown, T, Billeter, M, Lincoln, P, Norden, B, Albinsson, B, Wilhelmsson, L.M. | | Deposit date: | 2004-06-25 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA adopts normal B-form upon incorporation of highly fluorescent DNA base analogue tC: NMR structure and UV-Vis spectroscopy characterization.
Nucleic Acids Res., 32, 2004
|
|
1XYJ
 
 | | NMR Structure of the cat prion protein | | Descriptor: | prion protein | | Authors: | Lysek, D.A, Schorn, C, Nivon, L.G, Esteve-Moya, V, Christen, B, Calzolai, L, von Schroetter, C, Fiorito, F, Herrmann, T, Guntert, P. | | Deposit date: | 2004-11-10 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1U3M
 
 | | NMR structure of the chicken prion protein fragment 128-242 | | Descriptor: | prion-like protein | | Authors: | Lysek, D.A, Calzolai, L, Guntert, P, Wuthrich, K. | | Deposit date: | 2004-07-22 | | Release date: | 2005-01-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of chickens, turtles, and frogs
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1UR6
 
 | | NMR based structural model of the UbcH5B-CNOT4 complex | | Descriptor: | POTENTIAL TRANSCRIPTIONAL REPRESSOR NOT4HP, UBIQUITIN-CONJUGATING ENZYME E2-17 KDA 2, ZINC ION | | Authors: | Dominguez, C, Bonvin, A.M.J.J, Winkler, G.S, Van Schaik, F.M.A, Timmers, H.Th.M, Boelens, R. | | Deposit date: | 2003-10-27 | | Release date: | 2004-05-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Ubch5B/Cnot4 Complex Revealed by Combining NMR, Mutagenesis, and Docking Approaches.
Structure, 12, 2004
|
|
1UAP
 
 | | NMR structure of the NTR domain from human PCOLCE1 | | Descriptor: | Procollagen C-proteinase enhancer protein | | Authors: | Liepinsh, E, Banyai, L, Pintacuda, G, Trexler, M, Patthy, L, Otting, G. | | Deposit date: | 2003-03-14 | | Release date: | 2003-07-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Netrin-like Domain (NTR) of Human Type I Procollagen C-Proteinase Enhancer Defines Structural Consensus of NTR Domains and Assesses Potential Proteinase Inhibitory Activity and Ligand Binding.
J.Biol.Chem., 278, 2003
|
|
2MBB
 
 | | Solution Structure of the human Polymerase iota UBM1-Ubiquitin Complex | | Descriptor: | Immunoglobulin G-binding protein G/DNA polymerase iota fusion protein, Polyubiquitin-B | | Authors: | Wang, S, Zhou, P. | | Deposit date: | 2013-07-29 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparsely-sampled, high-resolution 4-D omit spectra for detection and assignment of intermolecular NOEs of protein complexes.
J.Biomol.Nmr, 59, 2014
|
|
2FNB
 
 | | NMR STRUCTURE OF THE FIBRONECTIN ED-B DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Fattorusso, R, Pellecchia, M, Viti, F, Neri, P, Neri, D, Wuthrich, K. | | Deposit date: | 1998-12-16 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the human oncofoetal fibronectin ED-B domain, a specific marker for angiogenesis.
Structure Fold.Des., 7, 1999
|
|
1TFS
 
 | | NMR AND RESTRAINED MOLECULAR DYNAMICS STUDY OF THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF TOXIN FS2, A SPECIFIC BLOCKER OF THE L-TYPE CALCIUM CHANNEL, ISOLATED FROM BLACK MAMBA VENOM | | Descriptor: | TOXIN FS2 | | Authors: | Albrand, J.-P, Blackledge, M.J, Pascaud, F, Hollecker, M, Marion, D. | | Deposit date: | 1995-01-26 | | Release date: | 1995-03-31 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR and restrained molecular dynamics study of the three-dimensional solution structure of toxin FS2, a specific blocker of the L-type calcium channel, isolated from black mamba venom.
Biochemistry, 34, 1995
|
|
2MOZ
 
 | | Structure of the Membrane Protein MerF, a Bacterial Mercury Transporter, Improved by the Inclusion of Chemical Shift Anisotropy Constraints | | Descriptor: | MerF | | Authors: | Tian, Y, Lu, G.J, Marassi, F.M, Opella, S.J, Membrane Protein Structures by Solution NMR (MPSbyNMR) | | Deposit date: | 2014-05-07 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane protein MerF, a bacterial mercury transporter, improved by the inclusion of chemical shift anisotropy constraints.
J.Biomol.Nmr, 60, 2014
|
|
1XY8
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
1XY4
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
1XYK
 
 | | NMR Structure of the canine prion protein | | Descriptor: | prion protein | | Authors: | Lysek, D.A, Schorn, C, Esteve-Moya, V, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-11-10 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1XYQ
 
 | | NMR structure of the pig prion protein | | Descriptor: | Major prion protein | | Authors: | Lysek, D.A, Schorn, C, Herrmann, T, Wuthrich, K. | | Deposit date: | 2004-11-10 | | Release date: | 2005-01-04 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of cats, dogs, pigs, and sheep
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1XY9
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
1XY5
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
1XY6
 
 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
2FQ8
 
 | |
2FQ5
 
 | |
1K6X
 
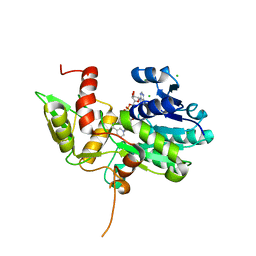 | | Crystal structure of Nmra, a negative transcriptional regulator in complex with NAD at 1.5 A resolution (Trigonal form) | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2002
|
|
1K6J
 
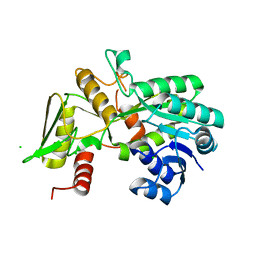 | | Crystal structure of Nmra, a negative transcriptional regulator (Monoclinic form) | | Descriptor: | CHLORIDE ION, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-16 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2001
|
|
1K6I
 
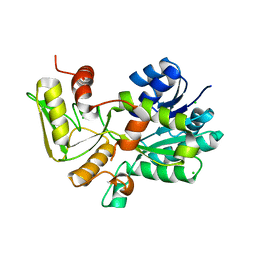 | | Crystal structure of Nmra, a negative transcriptional regulator (Trigonal form) | | Descriptor: | CHLORIDE ION, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-16 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2001
|
|
1WNN
 
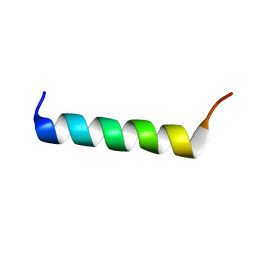 | | NMR structure of fmbp-1 tandem repeat 4 in 30%(v/v) TFE solution | | Descriptor: | FIBROIN-MODULATOR BINDING-PROTEIN-1 | | Authors: | Yamaki, T, Kawaguchi, K, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-08-06 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of FMBP-1 tandem repeat 4 in 30%(v/v) TFE solution
To be Published
|
|
1WNM
 
 | | NMR structure of FMBP-1 tandem repeat 2 in 30%(v/v) TFE solution | | Descriptor: | FIBROIN-MODULATOR BINDING-PROTEIN-1 | | Authors: | Yamaki, T, Kawaguchi, K, Aizawa, T, Takiya, S, Demura, M, Nitta, K. | | Deposit date: | 2004-08-06 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of FMBP-1 tandem repeat 2 in 30%(v/v) TFE solution
To be Published
|
|
2GBH
 
 | | NMR structure of stem region of helix-35 of 23S E.coli ribosomal RNA (residues 736-760) | | Descriptor: | 5'-R(*(GMP)P*GP*GP*CP*UP*AP*AP*UP*GP*(PSU)P*UP*GP*AP*AP*AP*AP*AP*UP*UP*AP*GP*CP*CP*C)-3' | | Authors: | Bax, A, Boisbouvier, J, Bryce, D, Grishaev, A, Jaroniec, C, Miclet, E, Nikonovicz, E, O'Neil-Cabello, E, Ying, J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics.
J.Am.Chem.Soc., 126, 2004
|
|
2GAQ
 
 | |
