2MMB
 
 | |
8IGD
 
 | |
1OL1
 
 | | Cyclin A binding groove inhibitor H-Cit-Cit-Leu-Ile-(p-F-Phe)-NH2 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CIR-CIR-LEU-ILE-PFF-NH2, CYCLIN A2 | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-08-04 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Cyclin Groove Recognition. Complex Crystal Structures and Inhibitor Design Through Ligand Exchange
Structure, 11, 2003
|
|
1OIY
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 4-(6-CYCLOHEXYLMETHOXY-9H-PURIN-2-YLAMINO)--BENZAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
3WJB
 
 | | Crystal structure of mutant nitrobindin M75L/H76L/Q96C/M148L/H158L (NB4) from Arabidopsis thaliana | | Descriptor: | BARIUM ION, HEXAETHYLENE GLYCOL, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Mizohata, E, Fukumoto, K, Onoda, A, Bocola, M, Arlt, M, Inoue, T, Schwaneberg, U, Hayashi, T. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Rhodium Complex-linked Hybrid Biocatalyst: Stereo-controlled Phenylacetylene Polymerization within an Engineered Protein Cavity
CHEMCATCHEM, 2014
|
|
3WBX
 
 | | Crystal structure of Gox0644 at apoform | | Descriptor: | Putative 2,5-diketo-D-gluconic acid reductase, SULFATE ION | | Authors: | Yuan, Y.A, Wang, C. | | Deposit date: | 2013-05-22 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Gox0644 at apoform
TO BE PUBLISHED
|
|
3WZK
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH N-cyclopropyl-4-(8-((thiophen-2-ylmethyl)amino)imidazo[1,2-a]pyrazin-3-yl)benzamide | | Descriptor: | CHLORIDE ION, Dual specificity protein kinase TTK, N-cyclopropyl-4-{8-[(thiophen-2-ylmethyl)amino]imidazo[1,2-a]pyrazin-3-yl}benzamide | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Mura, H, Nakamura, Y. | | Deposit date: | 2014-09-29 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of imidazo[1,2-b]pyridazine derivatives: selective and orally available Mps1 (TTK) kinase inhibitors exhibiting remarkable antiproliferative activity.
J.Med.Chem., 58, 2015
|
|
3WCQ
 
 | | Crystal structure analysis of Cyanidioschyzon melorae ferredoxin D58N mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Ueno, Y, Matsumoto, T, Yamano, A, Imai, T, Morimoto, Y. | | Deposit date: | 2013-05-31 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Increasing the electron-transfer ability of Cyanidioschyzon merolae ferredoxin by a one-point mutation - A high resolution and Fe-SAD phasing crystal structure analysis of the Asp58Asn mutant
Biochem.Biophys.Res.Commun., 436, 2013
|
|
1PCJ
 
 | |
3WYY
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH (E)-3-(4-((6-(((3s,5s,7s)-adamantan-1-yl)amino)-4-amino-5-cyanopyridin-2-yl)amino)-2-(cyanomethoxy)phenyl)-N-(2-methoxyethyl)acrylamide | | Descriptor: | (2E)-3-[4-({4-amino-5-cyano-6-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]dec-1-ylamino]pyridin-2-yl}amino)-2-(cyanomethoxy)phenyl]-N-(2-methoxyethyl)prop-2-enamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Yamamoto, T, Kojima, E, Mitsuoka, Y, Tadano, G, Tagashira, S, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Ando, S, Maeda, M, Higaki, M, Yoshizawa, H, Murai, H, Nakamura, Y. | | Deposit date: | 2014-09-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A unique hinge binder of extremely selective aminopyridine-based Mps1 (TTK) kinase inhibitors with cellular activity.
Bioorg.Med.Chem., 23, 2015
|
|
3WZH
 
 | | Crystal structure of AfCsx3 | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein AF_1864 | | Authors: | Yuan, Y.A, Yan, X. | | Deposit date: | 2014-09-25 | | Release date: | 2015-07-15 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease.
Rna Biol., 12, 2015
|
|
1O00
 
 | |
3ZQ9
 
 | | Structure of a Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44 | | Descriptor: | (2R,3S,4R,5R)-5-(HYDROXYMETHYL)PIPERIDINE-2,3,4-TRIOL, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
3ZS1
 
 | | Human Myeloperoxidase inactivated by TX5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-METHOXYETHYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
3ZO8
 
 | | Wild-type chorismate mutase of Bacillus subtilis at 1.6 A resolution | | Descriptor: | CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-20 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZU5
 
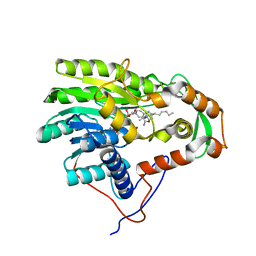 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT173 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(3-amino-2-methylbenzyl)-4-hexylpyridin-2(1H)-one, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3UUM
 
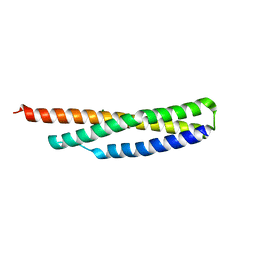 | |
3UWB
 
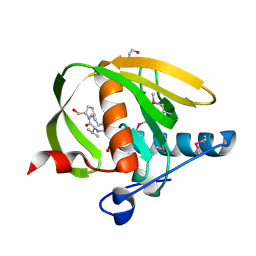 | | Crystal structure of a probable peptide deformylase from strucynechococcus phage S-SSM7 in complex with actinonin | | Descriptor: | 1,2-ETHANEDIOL, ACTINONIN, CHLORIDE ION, ... | | Authors: | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2011-12-01 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
3UDJ
 
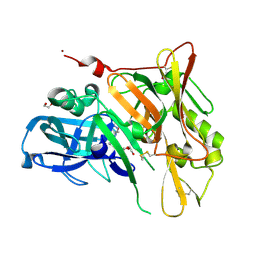 | | Crystal Structure of BACE with Compound 5 | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-18 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
3UWW
 
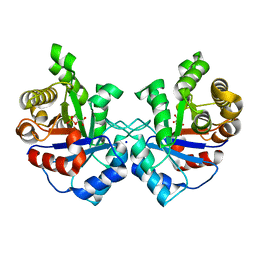 | | Crystal structure of Staphylococcus Aureus triosephosphate isomerase complexed with 3-phosphoglyceric acid | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-PHOSPHOGLYCERIC ACID, SODIUM ION, ... | | Authors: | Mukherjee, S, Roychowdhury, A, Dutta, D, Das, A.K. | | Deposit date: | 2011-12-03 | | Release date: | 2012-10-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
3ZOP
 
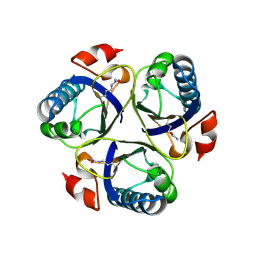 | | Arg90Cit chorismate mutase of Bacillus subtilis at 1.6 A resolution | | Descriptor: | CHORISMATE MUTASE AROH | | Authors: | Burschowsky, D, vanEerde, A, Okvist, M, Kienhofer, A, Kast, P, Hilvert, D, Krengel, U. | | Deposit date: | 2013-02-22 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Electrostatic Transition State Stabilization Rather Than Reactant Destabilization Provides the Chemical Basis for Efficient Chorismate Mutase Catalysis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ZQR
 
 | | NMePheB25 insulin analogue crystal structure | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
3ZD9
 
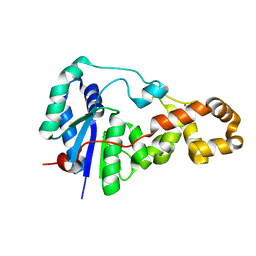 | | Potassium bound structure of E. coli ExoIX in P21 | | Descriptor: | POTASSIUM ION, PROTEIN XNI | | Authors: | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
3ZMM
 
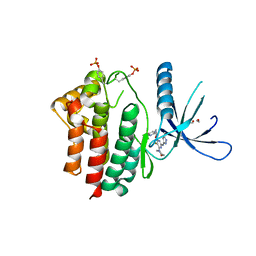 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | 5-FLUORO-4-[(1S)-1-(5-FLUOROPYRIMIDIN-2-YL)ETHOXY]-N-(5-METHYL-1H-PYRAZOL-3-YL)-6-MORPHOLINO-PYRIMIDIN-2-AMINE, ACETYL GROUP, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J, Green, I, Pollard, H, Howard, T, Mott, R. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Novel Jak2-Stat Pathway Inhibitors with Extended Residence Time on Target.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3WUC
 
 | | X-ray crystal structure of Xenopus laevis galectin-Va | | Descriptor: | Galectin, MALONIC ACID, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Nonaka, Y, Yoshida, H, Kamitori, S, Nakamura, T. | | Deposit date: | 2014-04-23 | | Release date: | 2015-04-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a Xenopus laevis skin proto-type galectin, close to but distinct from galectin-1.
Glycobiology, 25, 2015
|
|
