8BK5
 
 | | A structure of the truncated LpMIP with bound inhibitor JK095. | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase, SODIUM ION | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
5N4Z
 
 | |
7ADR
 
 | | CO bound as bridging ligand at the active site of vanadium nitrogenase VFe protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ... | | Authors: | Rohde, M, Grunau, K, Einsle, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | CO Binding to the FeV Cofactor of CO-Reducing Vanadium Nitrogenase at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8VJ2
 
 | |
8Q69
 
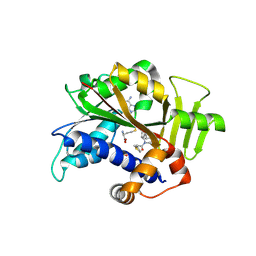 | | Crystal structure of HsRNMT complexed with inhibitor DDD1060606 | | Descriptor: | 1-[(3~{S})-3-(2~{H}-pyrazolo[3,4-b]pyridin-3-yl)piperidin-1-yl]-2-thiophen-3-yl-ethanone, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Petit, A.P, Fyfe, P.K. | | Deposit date: | 2023-08-11 | | Release date: | 2024-08-21 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Characterisation of RNA guanine-7 methyltransferase (RNMT) using a small molecule approach.
Biochem.J., 482, 2025
|
|
8RF2
 
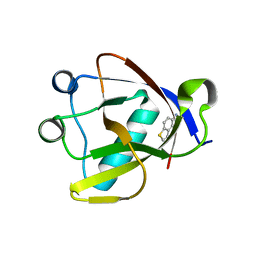 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 1E7 refined against the anomalous diffraction data | | Descriptor: | 1-benzothiophen-5-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8QG5
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-[6-azanyl-9-(phenylmethyl)purin-8-yl]prop-2-ynyl-methyl-amino]methyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To be published
|
|
8QG6
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[3-[6-azanyl-9-(phenylmethyl)purin-8-yl]prop-2-ynoxymethyl]oxolane-3,4-diol, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.142 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To be published
|
|
5MQK
 
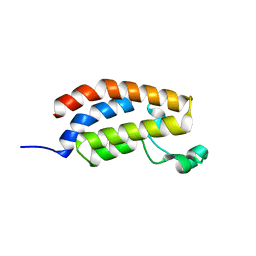 | |
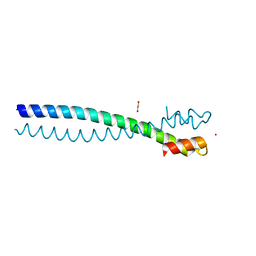
5JVS
 
 | |
5JVU
 
 | |
8EWU
 
 | | X-ray structure of the GDP-6-deoxy-4-keto-D-lyxo-heptose-4-reductase from Campylobacter jejuni HS:15 | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Riegert, A.S, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bifunctional Epimerase/Reductase Enzymes Facilitate the Modulation of 6-Deoxy-Heptoses Found in the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 62, 2023
|
|
6FTP
 
 | |
7ZZE
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},25~{R},26~{S},27~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-26,27-bis(oxidanyl)-28-oxa-2,4,6,9,14,17,21,23-octazatetracyclo[23.2.1.0^{2,10}.0^{3,8}]octacosa-3(8),4,6,9-tetraen-11-yne-16,22-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
7ZZF
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},23~{R},24~{S},25~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-24,25-bis(oxidanyl)-26-oxa-2,4,6,9,14,17,21-heptazatetracyclo[21.2.1.0^{2,10}.0^{3,8}]hexacosa-3(8),4,6,9-tetraen-11-yne-18,20-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
8TD7
 
 | | Structure of PYCR1 complexed with 2S-hydroxy-3-methylbutyric acid | | Descriptor: | (2S)-2-hydroxy-3-methylbutanoic acid, DI(HYDROXYETHYL)ETHER, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1).
Protein Sci., 33, 2024
|
|
6QAN
 
 | |
8HRQ
 
 | |
8ZJ5
 
 | | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3,4-Dihydroxybenzoate. | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Kumar, K.A, Pahwa, D, Kumar, P. | | Deposit date: | 2024-05-14 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3,4-Dihydroxybenzoate.
To Be Published
|
|
8ZJ0
 
 | | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3-Hydroxybenzoate. | | Descriptor: | 3-HYDROXYBENZOIC ACID, 4-hydroxythreonine-4-phosphate dehydrogenase, CHLORIDE ION, ... | | Authors: | Kumar, K.A, Pahwa, D, Kumar, P. | | Deposit date: | 2024-05-14 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3-Hydroxybenzoate.
To Be Published
|
|
8ZJ7
 
 | | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3,4-Dihydroxybenzoate and NAD. | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 4-hydroxythreonine-4-phosphate dehydrogenase, CHLORIDE ION, ... | | Authors: | Kumar, K.A, Pahwa, D, Kumar, P. | | Deposit date: | 2024-05-14 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Terephthalate 1,2-cis-dihydrodioldehydrogenase/Decarboxylase in complex with 3,4-Dihydroxybenzoate and NAD.
To Be Published
|
|
8RNP
 
 | |
6O04
 
 | | M.tb MenD IntII bound with Inhibitor | | Descriptor: | (1~{R},2~{S},5~{S},6~{S})-2-[(1~{S})-1-[3-[(4-azanylidene-2-methyl-1~{H}-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl (phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-3-ium-2-yl]-1,4-bis(oxidanyl)-4-oxidanylidene-butyl]-6-oxidanyl-5-(3-oxid anyl-3-oxidanylidene-prop-1-en-2-yl)oxy-cyclohex-3-ene-1-carboxylic acid, 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Johnston, J.M, Bashiri, G, Bulloch, E.M, Jirgis, E.M.N, Nigon, L.V, Chuang, H, Baker, E.N. | | Deposit date: | 2019-02-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric regulation of menaquinone (vitamin K2) biosynthesis in the human pathogenMycobacterium tuberculosis.
J.Biol.Chem., 295, 2020
|
|
9GV8
 
 | |
6NU1
 
 | | Crystal Structure of Human PKM2 in Complex with L-cysteine | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CYSTEINE, MAGNESIUM ION, ... | | Authors: | Srivastava, D, Nandi, S, Dey, M. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic and Structural Insights into Cysteine-Mediated Inhibition of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 58, 2019
|
|
