2D8Y
 
 | |
2KV1
 
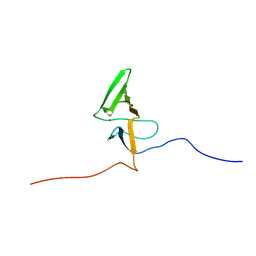 | | Insights into Function, Catalytic Mechanism and Fold Evolution of Mouse Selenoprotein Methionine Sulfoxide Reductase B1 through Structural Analysis | | Descriptor: | Methionine-R-sulfoxide reductase B1, ZINC ION | | Authors: | Aachmann, F.L, Sal, L.S, Kim, H.Y, Gladyshev, V.N, Dikiy, A. | | Deposit date: | 2010-03-04 | | Release date: | 2010-03-16 | | Last modified: | 2014-04-09 | | Method: | SOLUTION NMR | | Cite: | Insights into function, catalytic mechanism, and fold evolution of selenoprotein methionine sulfoxide reductase B1 through structural analysis
J.Biol.Chem., 285, 2010
|
|
2LHI
 
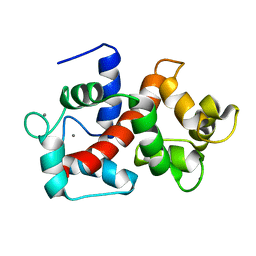 | | Solution structure of Ca2+/CNA1 peptide-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin,Serine/threonine-protein phosphatase 2B catalytic subunit A1 | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
2J1L
 
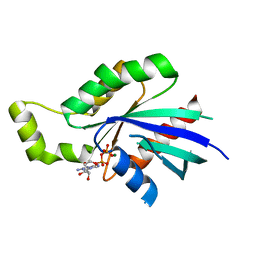 | | Crystal Structure of Human Rho-related GTP-binding protein RhoD | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RHO-RELATED GTP-BINDING PROTEIN RHOD | | Authors: | Pike, A.C.W, Johansson, C, Gileadi, C, Niesen, F.H, Sobott, F, Schoch, G, Elkins, J, Smee, C, Gorrec, F, Watt, S, Bray, J, Turnbull, A.P, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2006-08-14 | | Release date: | 2006-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Rho-Related GTP-Binding Protein Rhod
To be Published
|
|
2JW4
 
 | | NMR solution structure of the N-terminal SH3 domain of human Nckalpha | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Santiveri, C.M, Borroto, A, Simon, L, Rico, M, Ortiz, A.R, Alarcon, B, Jimenez, M. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Interaction between the N-terminal SH3 domain of Nckalpha and CD3epsilon-derived peptides: Non-canonical and canonical recognition motifs
BIOCHEM.BIOPHYS.ACTA PROTEINS & PROTEOMICS, 1794, 2009
|
|
1ISH
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with ethenoNADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ETHENO-NADP, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISF
 
 | | Crystal Structure Analysis of BST-1/CD157 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
2JXB
 
 | | Structure of CD3epsilon-Nck2 first SH3 domain complex | | Descriptor: | T-cell surface glycoprotein CD3 epsilon chain, Cytoplasmic protein NCK2 | | Authors: | Takeuchi, K, Yang, H, Ng, E, Park, S, Sun, Z.J, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-11-09 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional evidence that Nck interaction with CD3epsilon regulates T-cell receptor activity.
J.Mol.Biol., 380, 2008
|
|
2JS2
 
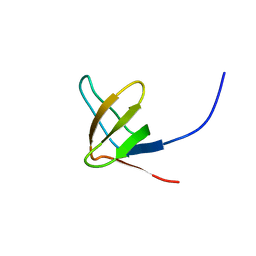 | |
1ISJ
 
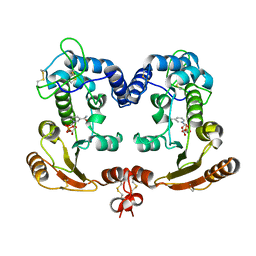 | | Crystal Structure Analysis of BST-1/CD157 complexed with NMN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISI
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with ethenoNAD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, [[(2R,3S,4R,5R)-3,4-dihydroxy-5-(9H-imidazo[2,1-f]purin-6-ium-3-yl)oxolan-2-yl]methoxy-oxidanidyl-phosphoryl] [(2R,3S,4R,5R)-3,4-dihydroxy-5-oxidanidyl-oxolan-2-yl]methyl phosphate, ... | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISG
 
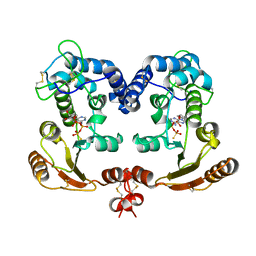 | | Crystal Structure Analysis of BST-1/CD157 with ATPgammaS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1ISM
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with nicotinamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
7LHT
 
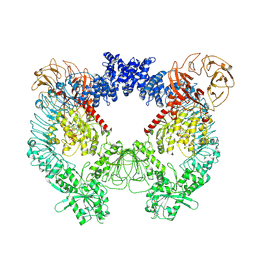 | | Structure of the LRRK2 dimer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LHW
 
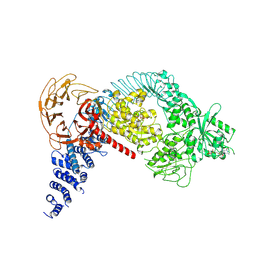 | | Structure of the LRRK2 monomer | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI3
 
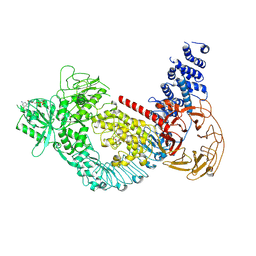 | | Structure of the LRRK2 G2019S mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
7LI4
 
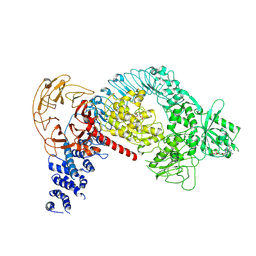 | | Structure of LRRK2 after symmetry expansion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Myasnikov, A, Zhu, H, Hixson, P, Xie, B, Yu, K, Pitre, A, Peng, J, Sun, J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural analysis of the full-length human LRRK2.
Cell, 184, 2021
|
|
4K17
 
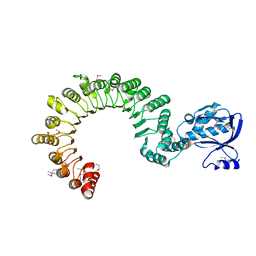 | | Crystal Structure of mouse CARMIL residues 1-668 | | Descriptor: | CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, Leucine-rich repeat-containing protein 16A, ... | | Authors: | Zwolak, A, Dominguez, R. | | Deposit date: | 2013-04-04 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | CARMIL leading edge localization depends on a non-canonical PH domain and dimerization.
Nat Commun, 4, 2013
|
|
4KAX
 
 | | Crystal structure of the Grp1 PH domain in complex with Arf6-GTP | | Descriptor: | ADP-ribosylation factor 6, CITRIC ACID, Cytohesin-3, ... | | Authors: | Lambright, D.G, Malaby, A.W, van den Berg, B. | | Deposit date: | 2013-04-23 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for membrane recruitment and allosteric activation of cytohesin family Arf GTPase exchange factors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7NA8
 
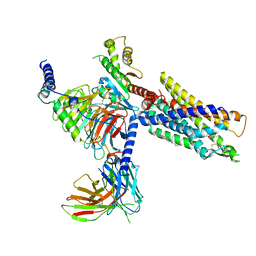 | | Structures of human ghrelin receptor-Gi complexes with ghrelin and a synthetic agonist | | Descriptor: | 1-(methanesulfonyl)-1'-(2-methyl-L-alanyl-O-benzyl-D-seryl)-1,2-dihydrospiro[indole-3,4'-piperidine], Antibody fragment, CHOLESTEROL, ... | | Authors: | Liu, H, Sun, D, Sun, J, Zhang, C. | | Deposit date: | 2021-06-20 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human ghrelin receptor signaling by ghrelin and the synthetic agonist ibutamoren
Nat Commun, 12, 2021
|
|
7NA7
 
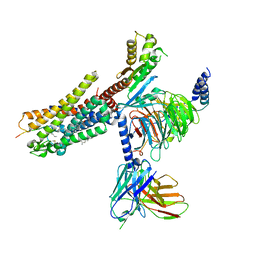 | | Structures of human ghrelin receptor-Gi complexes with ghrelin and a synthetic agonist | | Descriptor: | Antibody fragment, CHOLESTEROL, Ghrelin-27, ... | | Authors: | Liu, H, Sun, D, Sun, J, Zhang, C. | | Deposit date: | 2021-06-20 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of human ghrelin receptor signaling by ghrelin and the synthetic agonist ibutamoren
Nat Commun, 12, 2021
|
|
7NER
 
 | |
7NET
 
 | |
7NES
 
 | |
7NMZ
 
 | |
