3C7D
 
 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH-Pyruvate) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase, PYRUVIC ACID | | Authors: | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | Deposit date: | 2008-02-07 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
3C7C
 
 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH-L-Arginine) | | Descriptor: | ARGININE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | Deposit date: | 2008-02-07 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
2EYT
 
 | |
2F8O
 
 | |
2OO0
 
 | | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, ACETATE ION, Ornithine decarboxylase, ... | | Authors: | Dufe, V.T, Ingner, D, Khomutov, A.R, Heby, O, Persson, L, Al-Karadaghi, S. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane.
Biochem.J., 405, 2007
|
|
2J9Q
 
 | | A novel conformation for the TPR domain of pex5p | | Descriptor: | PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR, STRONTIUM ION | | Authors: | Stanley, W.A, Wilmanns, M, Kursula, P. | | Deposit date: | 2006-11-15 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Previously Unobserved Conformation for the Human Pex5P Receptor Suggests Roles for Intrinsic Flexibility and Rigid Domain Motions in Ligand Binding
Bmc Struct.Biol., 7, 2007
|
|
2BW7
 
 | | A novel mechanism for adenylyl cyclase inhibition from the crystal structure of its complex with catechol estrogen | | Descriptor: | 2,3,17BETA-TRIHYDROXY-1,3,5(10)-ESTRATRIENE, ADENYLATE CYCLASE, CALCIUM ION, ... | | Authors: | Steegborn, C, Litvin, T.N, Hess, K.C, Capper, A.B, Taussig, R, Buck, J, Levin, L.R, Wu, H. | | Deposit date: | 2005-07-12 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Novel Mechanism for Adenylyl Cyclase Inhibition from the Crystal Structure of its Complex with Catechol Estrogen
J.Biol.Chem., 280, 2005
|
|
3RTR
 
 | | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Calabrese, M.F, Scott, D.C, Duda, D.M, Grace, C.R, Kurinov, I, Kriwacki, R.W, Schulman, B.A. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1TNT
 
 | |
1TNS
 
 | |
2ON3
 
 | | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase | | Authors: | Dufe, V.T, Ingner, D, Heby, O, Khomutov, A.R, Persson, L, Al-Karadaghi, S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane.
Biochem.J., 405, 2007
|
|
2IBM
 
 | |
2JAB
 
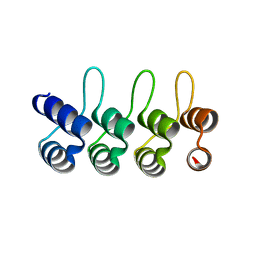 | | A designed ankyrin repeat protein evolved to picomolar affinity to Her2 | | Descriptor: | H10-2-G3 | | Authors: | Zahnd, C, Wyler, E, Schwenk, J.M, Steiner, D, Lawrence, M.C, McKern, N.M, Pecorari, F, Ward, C.W, Joos, T.O, Pluckthun, A. | | Deposit date: | 2006-11-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Designed Ankyrin Repeat Protein Evolved to Picomolar Affinity to Her2
J.Mol.Biol., 369, 2007
|
|
2EYS
 
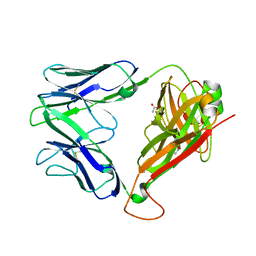 | |
2K0E
 
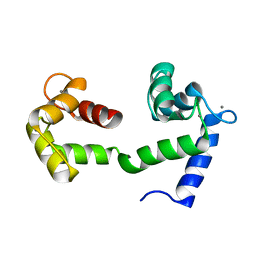 | | A Coupled Equilibrium Shift Mechanism in Calmodulin-Mediated Signal Transduction | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Gsponer, J, Christodoulou, J, Cavalli, A, Bui, J.M, Richter, B, Dobson, C.M, Vendruscolo, M. | | Deposit date: | 2008-02-02 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A coupled equilibrium shift mechanism in calmodulin-mediated signal transduction
Structure, 16, 2008
|
|
2EYR
 
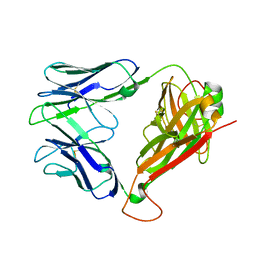 | |
2JB4
 
 | | Isopenicillin N synthase with a 2-thiabicycloheptan-6-one product analogue | | Descriptor: | (1S,4S,5S,7R)-7-{[(5S)-5-AMINO-5-CARBOXYPENTANOYL]AMINO}-3,3-DIMETHYL-6-OXO-2-THIABICYCLO[3.2.0]HEPTANE-4-CARBOXYLIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Stewart, A.C, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Cyclobutanone Analogue Mimics Penicillin in Binding to Isopenicillin N Synthase.
Chembiochem, 8, 2007
|
|
2LFM
 
 | |
2KJI
 
 | | A divergent ins protein in c. elegans structurally resemble insulin and activates the human insulin receptor | | Descriptor: | Probable insulin-like peptide beta-type 5 | | Authors: | Hua, Q.X, Nakarawa, S.H, Wilken, R, Ramos, R.R, Jia, W.H, Bass, J, Weiss, M.A. | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-16 | | Last modified: | 2019-09-18 | | Method: | SOLUTION NMR | | Cite: | A divergent INS protein in Caenorhabditis elegans structurally resembles human insulin and activates the human insulin receptor.
Genes Dev., 17, 2003
|
|
2MC6
 
 | | A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement | | Descriptor: | DNA-directed RNA polymerase subunit beta', RNA polymerase inhibitor p7 | | Authors: | Liu, B, Shadrin, A, Sheppard, C, Xu, Y, Severinov, K, Matthews, S, Wigneshweraraj, S. | | Deposit date: | 2013-08-15 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement.
Nucleic Acids Res., 42, 2014
|
|
2KQO
 
 | |
2MC5
 
 | | A bacteriophage transcription regulator inhibits bacterial transcription initiation by -factor displacement | | Descriptor: | 45L | | Authors: | Liu, B, Shadrin, A, Sheppard, C, Xu, Y, Severinov, K, Matthews, S, Wigneshweraraj, S. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A bacteriophage transcription regulator inhibits bacterial transcription initiation by sigma-factor displacement.
Nucleic Acids Res., 42, 2014
|
|
7NAS
 
 | | Bacterial 30S ribosomal subunit assembly complex state A (multibody refinement for body domain of 30S ribosome) | | Descriptor: | 16S rRNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
6XCB
 
 | |
6XCC
 
 | |
