3UQY
 
 | | H2-reduced structure of E. coli hydrogenase-1 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3USE
 
 | | Crystal Structure of E. coli hydrogenase-1 in its as-isolated form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1HPV
 
 | |
1HVI
 
 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2R,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
5FZC
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment 4,5-dihydronaphtho(1,2-b)thiophene-2- carboxylicacid (N11181a) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment 4,5-Dihydronaphtho(1,2-B)Thiophene-2-Carboxylicacid (N11181A) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
4LII
 
 | |
1ODX
 
 | | HIV-1 Proteinase mutant A71T, V82A | | Descriptor: | HIV-1 PROTEASE, di-tert-butyl {iminobis[(2S,3S)-3-hydroxy-1-phenylbutane-4,2-diyl]}biscarbamate | | Authors: | Kervinen, J, Thanki, N, Zdanov, A, Wlodawer, A. | | Deposit date: | 1996-09-16 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of the Native and Drug-Resistant HIV-1 Proteinases Complexed with an Aminodiol Inhibitor
Protein Pept.Lett., 3, 1996
|
|
1ODW
 
 | | Native HIV-1 Proteinase | | Descriptor: | HIV-1 PROTEASE, di-tert-butyl {iminobis[(2S,3S)-3-hydroxy-1-phenylbutane-4,2-diyl]}biscarbamate | | Authors: | Thanki, N, Kervinen, J, Wlodawer, A. | | Deposit date: | 1996-09-16 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of the Native and Drug-Resistant HIV-1 Proteinases Complexed with an Aminodiol Inhibitor
Protein Pept.Lett., 3, 1996
|
|
1HVJ
 
 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-3-HYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
1HVK
 
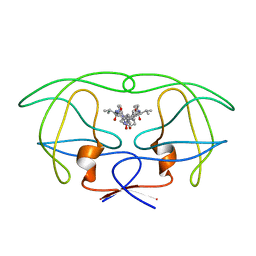 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2S,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
1HVL
 
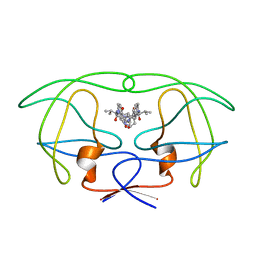 | | INFLUENCE OF STEREOCHEMISTRY ON ACTIVITY AND BINDING MODES FOR C2 SYMMETRY-BASED DIOL INHIBITORS OF HIV-1 PROTEASE | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2R,3R)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Hosur, M.V, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-01-26 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influence of Stereochemistry on Activity and Binding Modes for C2 Symmetry-Based Diol Inhibitors of HIV-1 Protease
J.Am.Chem.Soc., 116, 1994
|
|
4FAG
 
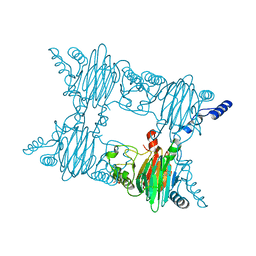 | | Crystal Structure of the Salicylate 1,2-dioxygenase from Pseudoaminobacter salicylatoxidans W104Y mutant in complex with gentisate | | Descriptor: | 2,5-dihydroxybenzoic acid, FE (III) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2012-05-22 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The generation of a 1-hydroxy-2-naphthoate 1,2-dioxygenase by single point mutations of salicylate 1,2-dioxygenase - Rational design of mutants and the crystal structures of the A85H and W104Y variants.
J.Struct.Biol., 180, 2012
|
|
3NJZ
 
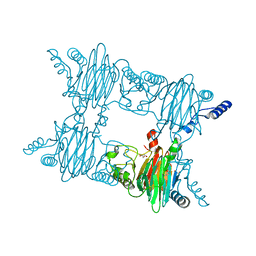 | | Crystal Structure of Salicylate 1,2-dioxygenase from Pseudoaminobacter salicylatoxidans Adducts with salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-06-18 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: A step towards the comprehension of the structural basis for substrate selection in class III ring cleaving dioxygenases.
J.Struct.Biol., 177, 2012
|
|
3NL1
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase from Pseudoaminobacter salicylatoxidans Adducts with gentisate | | Descriptor: | 2,5-dihydroxybenzoic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: A step towards the comprehension of the structural basis for substrate selection in class III ring cleaving dioxygenases.
J.Struct.Biol., 177, 2012
|
|
1MTR
 
 | | HIV-1 PROTEASE COMPLEXED WITH A CYCLIC PHE-ILE-VAL PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, SULFATE ION, [1-BENZYL-3-(8-SEC-BUTYL-7,10-DIOXO-2-OXA-6,9-DIAZA-BICYCLO[11.2.2] HEPTADECA-1(16),13(17),14-TRIEN-11-YLAMINO)-2-HYDROXY-PROPYL]-CARBAMIC ACID TERT-BUTYL ESTER | | Authors: | Wickramasinghe, W, Begun, J, Martin, J.L. | | Deposit date: | 1996-02-15 | | Release date: | 1996-08-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Substrate-based cyclic peptidomimetics of Phe-Ile-Val that inhibit HIV-1 protease using a novel enzyme-binding mode.
J.Am.Chem.Soc., 118, 1996
|
|
3NST
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase G106A mutant from Pseudoaminobacter salicylatoxidans | | Descriptor: | FE (II) ION, GLYCEROL, Gentisate 1,2-dioxygenase | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-07-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate.
FEBS J., 280, 2013
|
|
3KCX
 
 | | Factor inhibiting HIF-1 alpha in complex with Clioquinol | | Descriptor: | 5-chloro-7-iodoquinolin-8-ol, FE (II) ION, GLYCEROL, ... | | Authors: | Moon, H, Han, S, Choe, J. | | Deposit date: | 2009-10-22 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human FIH-1 in complex with quinol family inhibitors
Mol.Cells, 29, 2010
|
|
4JZR
 
 | | Structure of Prolyl Hydroxylase Domain-containing Protein (PHD) with Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 2-(biphenyl-4-yl)-8-[(1-methyl-1H-imidazol-2-yl)methyl]-2,8-diazaspiro[4.5]decan-1-one, Egl nine homolog 1, ... | | Authors: | Ma, Y, Yang, L. | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel complex crystal structure of prolyl hydroxylase domain-containing protein 2 (PHD2): 2,8-Diazaspiro[4.5]decan-1-ones as potent, orally bioavailable PHD2 inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
3HOK
 
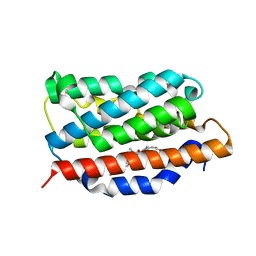 | | X-ray Crystal Structure of Human Heme Oxygenase-1 with (2R, 4S)-2-[2-(4-Chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-4[((5-trifluoromethylpyridin-2-yl)thio)methyl]-1,3-dioxolane: A Novel, Inducible Binding Mode | | Descriptor: | 2-({[(2R,4S)-2-[2-(4-chlorophenyl)ethyl]-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methyl}sulfanyl)-5-(trifluoromethyl)pyridine, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rahman, M.N, Jia, Z. | | Deposit date: | 2009-06-02 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | X-ray crystal structure of human heme oxygenase-1 with (2R,4S)-2-[2-(4-chlorophenyl)ethyl]-2-[(1H-imidazol-1-yl)methyl]-4[((5-trifluoromethylpyridin-2-yl)thio)methyl]-1,3-dioxolane: a novel, inducible binding mode.
J.Med.Chem., 52, 2009
|
|
1KBQ
 
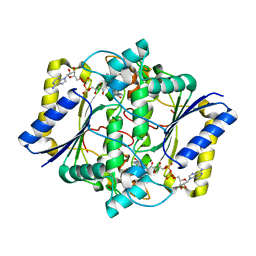 | | Complex of Human NAD(P)H quinone Oxidoreductase with 5-methoxy-1,2-dimethyl-3-(4-nitrophenoxymethyl)indole-4,7-dione (ES936) | | Descriptor: | 5-METHOXY-1,2-DIMETHYL-3-(4-NITROPHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Faig, M, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
1KBO
 
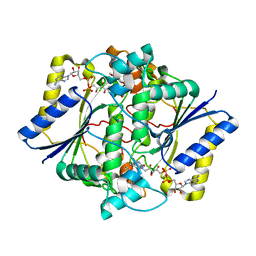 | | Complex of Human recombinant NAD(P)H:Quinone Oxide reductase type 1 with 5-methoxy-1,2-dimethyl-3-(phenoxymethyl)indole-4,7-dione (ES1340) | | Descriptor: | 5-METHOXY-1,2-DIMETHYL-3-(PHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Faig, M, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
1KNF
 
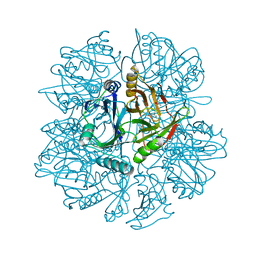 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with 3-methyl Catechol under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, 3-METHYLCATECHOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
3NVC
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase G106A mutant from Pseudoaminobacter salicylatoxidans in complex with salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate.
FEBS J., 280, 2013
|
|
3NW4
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase G106A mutant from Pseudoaminobacter salicylatoxidans in complex with gentisate | | Descriptor: | 2,5-dihydroxybenzoic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-07-09 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The salicylate 1,2-dioxygenase as a model for a conventional gentisate 1,2-dioxygenase: crystal structures of the G106A mutant and its adducts with gentisate and salicylate.
FEBS J., 280, 2013
|
|
4HM2
 
 | | Naphthalene 1,2-Dioxygenase bound to ethylphenylsulfide | | Descriptor: | (ethylsulfanyl)benzene, 1,2-ETHANEDIOL, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | One enzyme, many reactions: structural basis for the various reactions catalyzed by naphthalene 1,2-dioxygenase.
Iucrj, 4, 2017
|
|
