1PX8
 
 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
4MUT
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Alanine | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, D,D-dipeptidase/D,D-carboxypeptidase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3SS7
 
 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.55 A resolution | | Descriptor: | D-serine dehydratase, GLYCEROL, POTASSIUM ION, ... | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-01-18 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
4UMD
 
 | |
4UM5
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Moraxella catarrhalis in complex with Magnesium ion and Phosphate ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION, ... | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UM7
 
 | | Crystal structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase (kdsC) from Moraxella catarrhalis in complex with Magnesium ion | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-MANNO-OCTULOSONATE 8-PHOSPHATE PHOSPHATASE KDSC, MAGNESIUM ION | | Authors: | Dhindwal, S, Tomar, S, Kumar, P. | | Deposit date: | 2014-05-15 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ligand-Bound Structures of 3-Deoxy-D-Manno-Octulosonate 8-Phosphate Phosphatase from Moraxella Catarrhalis Reveal a Water Channel Connecting to the Active Site for the Second Step of Catalysis
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7OSE
 
 | | cytochrome bd-II type oxidase with bound aurachin D | | Descriptor: | Aurachin D, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-II ubiquinol oxidase subunit 1, ... | | Authors: | Grauel, A, Kaegi, J, Rasmussen, T, Wohlwend, D, Boettcher, B, Friedrich, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Escherichia coli cytochrome bd-II type oxidase with bound aurachin D.
Nat Commun, 12, 2021
|
|
6YYI
 
 | | Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase, CITRIC ACID, ... | | Authors: | Lafite, P, Daniellou, R, Bretagne, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of Dictyoglomus thermophilum beta-d-xylosidase DtXyl unravels the structural determinants for efficient notoginsenoside R1 hydrolysis.
Biochimie, 181, 2020
|
|
5F24
 
 | | Crystal structure of dual specific IMPase/NADP phosphatase bound with D-inositol-1-phosphate | | Descriptor: | CALCIUM ION, CHLORIDE ION, D-MYO-INOSITOL-1-PHOSPHATE, ... | | Authors: | Bhattacharyya, S, Dutta, D, Ghosh, A.K, Das, A.K. | | Deposit date: | 2015-12-01 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural elucidation of the NADP(H) phosphatase activity of staphylococcal dual-specific IMPase/NADP(H) phosphatase
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1KOJ
 
 | | Crystal structure of rabbit phosphoglucose isomerase complexed with 5-phospho-D-arabinonohydroxamic acid | | Descriptor: | 5-PHOSPHO-D-ARABINOHYDROXAMIC ACID, Glucose-6-phosphate isomerase | | Authors: | Arsenieva, D, Hardre, R, Salmon, L, Jeffery, C.J. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of rabbit phosphoglucose isomerase complexed with 5-phospho-D-arabinonohydroxamic acid.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6HZZ
 
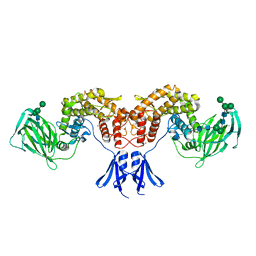 | | Structure of human D-glucuronyl C5 epimerase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8XIA
 
 | |
6I01
 
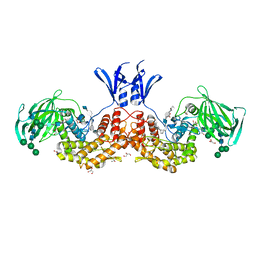 | | Structure of human D-glucuronyl C5 epimerase in complex with substrate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6I02
 
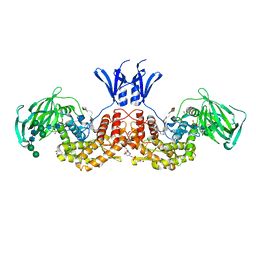 | | Structure of human D-glucuronyl C5 epimerase in complex with product | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Debarnot, C, Monneau, Y.R, Roig-Zamboni, V, Le Narvor, C, Goulet, A, Fadel, F, Vives, R.R, Bonnaffe, D, Lortat-Jacob, H, Bourne, Y. | | Deposit date: | 2018-10-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1KNJ
 
 | | Co-Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli Involved in Mevalonate-Independent Isoprenoid Biosynthesis, Complexed with CMP/MECDP/Mn2+ | | Descriptor: | 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE, 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
5A3O
 
 | | Crystal structure of the LecB lectin from Pseudomonas aeruginosa in complex with Methyl 6-(cinnamido)-6-deoxy-alpha-D-mannopyranoside at 1.6 ansgtrom | | Descriptor: | CALCIUM ION, CHLORIDE ION, CINNAMIDE, ... | | Authors: | Sommer, R, Hauck, D, Varrot, A, Audfray, A, Imberty, A, Titz, A. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cinnamide Derivatives of D-Mannose as Inhibitors of the Bacterial Virulence Factor Lecb from Pseudomonas Aeruginosa
Chemistryopen, 4, 2015
|
|
1JMA
 
 | | CRYSTAL STRUCTURE OF THE HERPES SIMPLEX VIRUS GLYCOPROTEIN D BOUND TO THE CELLULAR RECEPTOR HVEA/HVEM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCOPROTEIN D, HERPESVIRUS ENTRY MEDIATOR, ... | | Authors: | Carfi, A, Willis, S.H, Whitbeck, J.C, Krummenacker, C, Cohen, G.H, Eisenberg, R.J, Wiley, D.C. | | Deposit date: | 2001-07-17 | | Release date: | 2001-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Herpes simplex virus glycoprotein D bound to the human receptor HveA.
Mol.Cell, 8, 2001
|
|
5HHC
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21/n | | Descriptor: | D- Vascular endothelial growth factor-A, GLYCEROL, Vascular endothelial growth factor A | | Authors: | Uppalapati, M, Lee, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
5HHD
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {RFX037 plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D-Peptide RFX037.D, D-Vascular endothelial growth factor, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Uppalapati, M, LEE, D.J, Mandal, K, Kent, S.B.H, Sidhu, S. | | Deposit date: | 2016-01-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent d-Protein Antagonist of VEGF-A is Nonimmunogenic, Metabolically Stable, and Longer-Circulating in Vivo.
Acs Chem.Biol., 11, 2016
|
|
344D
 
 | | DETERMINATION BY MAD-DM OF THE STRUCTURE OF THE DNA DUPLEX D(ACGTACG(5-BRU))2 AT 1.46A AND 100K | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*AP*CP*GP*(BRU))-3') | | Authors: | Todd, A.R, Adams, A, Powell, H.R, Cardin, C.J. | | Deposit date: | 1997-08-04 | | Release date: | 1997-09-26 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Determination by MAD-DM of the structure of the DNA duplex d[ACGTACG(5-BrU)]2 at 1.46 A and 100 K.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4AZ6
 
 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-23 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
1K06
 
 | | Crystallographic Binding Study of 100 mM N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b | | Descriptor: | Glycogen Phosphorylase, N-[(phenylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2001-09-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of N-acetyl-N '-beta-D-glucopyranosyl urea and N-benzoyl-N '-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
1KTI
 
 | | BINDING OF 100 MM N-ACETYL-N'-BETA-D-GLUCOPYRANOSYL UREA TO GLYCOGEN PHOSPHORYLASE B: KINETIC AND CRYSTALLOGRAPHIC STUDIES | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-(acetylcarbamoyl)-beta-D-glucopyranosylamine, ... | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2002-01-16 | | Release date: | 2002-01-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Binding of N-acetyl-N'-beta-D-glucopyranosyl urea and N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
1MYT
 
 | | CRYSTAL STRUCTURE TO 1.74 ANGSTROMS RESOLUTION OF METMYOGLOBIN FROM YELLOWFIN TUNA (THUNNUS ALBACARES): AN EXAMPLE OF A MYOGLOBIN LACKING THE D HELIX | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Birnbaum, G.I, Evans, S.V, Przybylska, M, Rose, D.R. | | Deposit date: | 1991-05-06 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | 1.70 A resolution structure of myoglobin from yellowfin tuna. An example of a myoglobin lacking the D helix.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
2VO0
 
 | | Structure of PKA-PKB chimera complexed with C-(4-(4-Chlorophenyl)-1-(7H-pyrrolo(2,3-d)pyrimidin-4-yl)piperidin-4-yl)methylamine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, 1-[4-(4-chlorophenyl)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-4-yl]methanamine, ... | | Authors: | Caldwell, J.J, Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Aherne, G.W, Hunter, L.K, Taylor, K, Ruddle, R, Raynaud, F.I, Verdonk, M, Workman, P, Garrett, M.D, Collins, I. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Identification of 4-(4-Aminopiperidin-1-Yl)-7H-Pyrrolo[2,3-D]Pyrimidines as Selective Inhibitors of Protein Kinase B Through Fragment Elaboration.
J.Med.Chem., 51, 2008
|
|
