3HX6
 
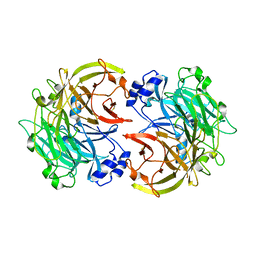 | |
1BF8
 
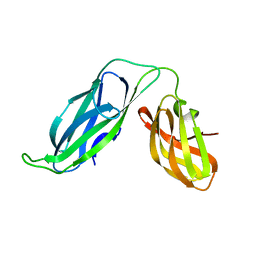 | | PERIPLASMIC CHAPERONE FIMC, NMR, 20 STRUCTURES | | Descriptor: | CHAPERONE PROTEIN FIMC | | Authors: | Pellecchia, M, Guntert, P, Glockshuber, R, Wuthrich, K. | | Deposit date: | 1998-05-28 | | Release date: | 1998-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the periplasmic chaperone FimC.
Nat.Struct.Biol., 5, 1998
|
|
3Q48
 
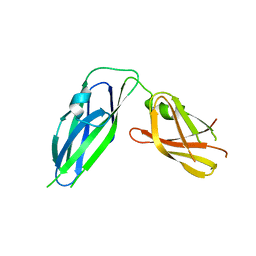 | | Crystal structure of Pseudomonas aeruginosa CupB2 chaperone | | Descriptor: | Chaperone CupB2 | | Authors: | Cai, X, Wang, R, Filloux, A, Waksman, G, Meng, G. | | Deposit date: | 2010-12-23 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional characterization of Pseudomonas aeruginosa CupB chaperones
Plos One, 6, 2011
|
|
3JWN
 
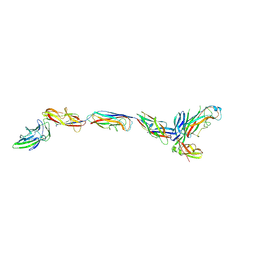 | | Complex of FimC, FimF, FimG and FimH | | Descriptor: | Chaperone protein fimC, FimH protein, GLYCEROL, ... | | Authors: | Le Trong, I, Aprikian, P, Stenkamp, R.E, Sokurenko, E.V. | | Deposit date: | 2009-09-18 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting.
Cell(Cambridge,Mass.), 141, 2010
|
|
5TC1
 
 | | In situ structures of the genome and genome-delivery apparatus in ssRNA bacteriophage MS2 | | Descriptor: | Capsid protein, Maturation protein, phage MS2 genome | | Authors: | Dai, X.H, Li, Z.H, Lai, M, Shu, S, Du, Y.S, Zhou, Z.H, Sun, R. | | Deposit date: | 2016-09-13 | | Release date: | 2016-12-07 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus.
Nature, 541, 2017
|
|
3BFW
 
 | |
3BFQ
 
 | |
1TR7
 
 | | FimH adhesin receptor binding domain from uropathogenic E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, FimH protein, ... | | Authors: | Bouckaert, J, Berglund, J, Schembri, M, De Genst, E, Cools, L, Wuhrer, M, Hung, C.S, Pinkner, J, Slattegard, R, Zavialov, A, Choudhury, D, Langermann, S, Hultgren, S.J, Wyns, L, Klemm, P, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-06-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin
Mol.Microbiol., 55, 2005
|
|
3OHN
 
 | | Crystal structure of the FimD translocation domain | | Descriptor: | Outer membrane usher protein FimD | | Authors: | Wang, T, Li, H. | | Deposit date: | 2010-08-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
5AFO
 
 | |
1UWF
 
 | | 1.7 A resolution structure of the receptor binding domain of the FimH adhesin from uropathogenic E. coli | | Descriptor: | FIMH PROTEIN, GLYCEROL, butyl alpha-D-mannopyranoside | | Authors: | Bouckaert, J, Berglund, J, Genst, E.D, Cools, L, Hung, C.-S, Wuhrer, M, Zavialov, A, Langermann, S, Hultgren, S, Wyns, L, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-02-05 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Receptor Binding Studies Disclose a Novel Class of High-Affinity Inhibitors of the Escherichia Coli Fimh Adhesin.
Mol.Microbiol., 55, 2005
|
|
4XO9
 
 | | Crystal structure of a FimH*DsG complex from E.coli K12 in space group C2 | | Descriptor: | Minor component of type 1 fimbriae, Protein FimH | | Authors: | Jakob, R.P, Eras, J, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
4XOD
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 | | Descriptor: | FimG protein, FimH protein | | Authors: | Jakob, R.P, Sauer, M.M, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
4XOA
 
 | | Crystal structure of a FimH*DsG complex from E.coli K12 in space group P1 | | Descriptor: | FimG, Protein FimH | | Authors: | Jakob, R.P, Eras, J, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.541 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
2KT6
 
 | | Structural homology between the C-terminal domain of the PapC usher and its plug | | Descriptor: | Outer membrane usher protein papC | | Authors: | Ford, B, Rego, A, Ragan, T.J, Pinkner, J, Dodson, K, Driscoll, P.C, Hultgren, S, Waksman, G. | | Deposit date: | 2010-01-19 | | Release date: | 2010-04-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural Homology between the C-Terminal Domain of the PapC Usher and Its Plug.
J.Bacteriol., 192, 2010
|
|
3L48
 
 | |
7QUO
 
 | | FimH lectin domain in complex with oligomannose-6 | | Descriptor: | FimH, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Bouckaert, J, Bourenkov, G.P. | | Deposit date: | 2022-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into a cooperative switch between one and two FimH bacterial adhesins binding pauci- and high-mannose type N-glycan receptors
J.Biol.Chem., 299, 2023
|
|
4XO8
 
 | | Crystal structure of the FimH lectin domain from E.coli K12 in complex with heptyl alpha-D-mannopyrannoside | | Descriptor: | Protein FimH, heptyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Eras, J, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
4XOC
 
 | | Crystal structure of the FimH lectin domain from E.coli F18 in complex with heptyl alpha-D-mannopyrannoside | | Descriptor: | FimH protein, heptyl alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Sauer, M.M, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
4XOB
 
 | | Crystal structure of a FimH*DsF complex from E.coli K12 with bound heptyl alpha-D-mannopyrannoside | | Descriptor: | FimF, Protein FimH, SULFATE ION, ... | | Authors: | Jakob, R.P, Eras, J, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
4XOE
 
 | | Crystal structure of a FimH*DsG complex from E.coli F18 with bound heptyl alpha-D-mannopyrannoside | | Descriptor: | CACODYLATE ION, FimG protein, FimH protein, ... | | Authors: | Jakob, R.P, Sauer, M.M, Navarra, G, Ernst, B, Glockshuber, R, Maier, T. | | Deposit date: | 2015-01-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catch-bond mechanism of the bacterial adhesin FimH.
Nat Commun, 7, 2016
|
|
2RJZ
 
 | | Crystal structure of the type 4 fimbrial biogenesis protein PilO from Pseudomonas aeruginosa | | Descriptor: | PilO protein, SULFATE ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex.
J.Mol.Biol., 394, 2009
|
|
3DPA
 
 | |
3TBE
 
 | |
2IVW
 
 | | The solution structure of a domain from the Neisseria meningitidis PilP pilot protein. | | Descriptor: | PILP PILOT PROTEIN | | Authors: | Golovanov, A.P, Balasingham, S, Tzitzilonis, C, Goult, B.T, Lian, L.-Y, Homberset, H, Tonjum, T, Derrick, J.P. | | Deposit date: | 2006-06-20 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a domain from the Neisseria meningitidis lipoprotein PilP reveals a new beta-sandwich fold.
J. Mol. Biol., 364, 2006
|
|
