2D3D
 
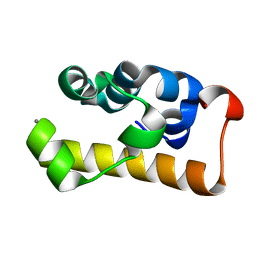 | | crystal structure of the RNA binding SAM domain of saccharomyces cerevisiae Vts1 | | Descriptor: | CALCIUM ION, Vts1 protein | | Authors: | Aviv, T, Amborski, A.N, Zhao, X.S, Kwan, J.J, Johnson, P.E, Sicheri, F, Donaldson, L.W. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The NMR and X-ray Structures of the Saccharomyces cerevisiae Vts1 SAM Domain Define a Surface for the Recognition of RNA Hairpins
J.Mol.Biol., 356, 2006
|
|
2FU0
 
 | | Plasmodium falciparum cyclophilin PFE0505w putative cyclosporin-binding domain | | Descriptor: | cyclophilin, putative | | Authors: | Dong, A, Lew, J, Sundararajan, E, Zhao, Y, Wasney, G, Vedadi, M, Koeieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-25 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
6NXA
 
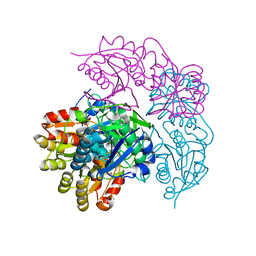 | | ECAII(D90T,K162T) MUTANT AT PH 7 | | Descriptor: | ACETIC ACID, GLYCEROL, L-asparaginase 2 | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2019-02-08 | | Release date: | 2019-08-07 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Opportunistic complexes of E. coli L-asparaginases with citrate anions.
Sci Rep, 9, 2019
|
|
6NX8
 
 | |
6O9Y
 
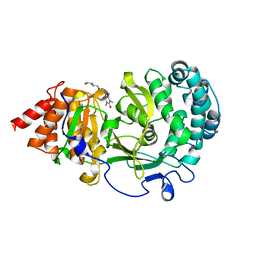 | | Structure of human PARG complexed with JA2-8 | | Descriptor: | 7-[(2S)-2-hydroxy-3-(morpholin-4-yl)propyl]-1,3-dimethyl-3,7-dihydro-1H-purine-2,6-dione, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
6OA3
 
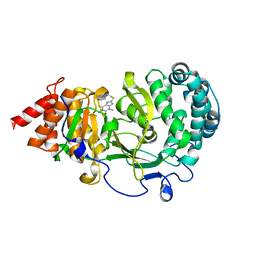 | | Structure of human PARG complexed with JA2131 | | Descriptor: | (8S)-1,3-dimethyl-8-{[2-(morpholin-4-yl)ethyl]sulfanyl}-6-sulfanylidene-1,3,6,7,8,9-hexahydro-2H-purin-2-one, Poly(ADP-ribose) glycohydrolase | | Authors: | Stegeman, R.A, Jones, D.E, Ellenberger, T, Kim, I.K, Tainer, J.A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selective small molecule PARG inhibitor causes replication fork stalling and cancer cell death.
Nat Commun, 10, 2019
|
|
4OTU
 
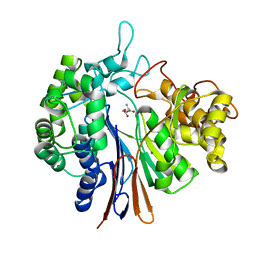 | |
6OYB
 
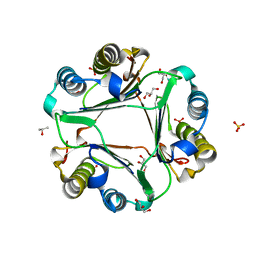 | |
6OII
 
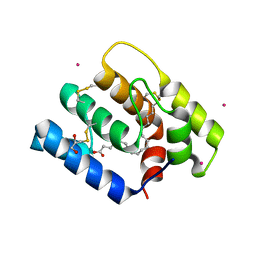 | |
1R70
 
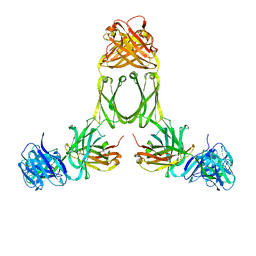 | | Model of human IgA2 determined by solution scattering, curve fitting and homology modelling | | Descriptor: | Human IgA2(m1) Heavy Chain, Human IgA2(m1) Light Chain | | Authors: | Furtado, P.B, Whitty, P.W, Robertson, A, Eaton, J.T, Almogren, A, Kerr, M.A, Woof, J.M, Perkins, S.J. | | Deposit date: | 2003-10-17 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | Solution Structure Determination of Monomeric Human IgA2 by X-ray and Neutron Scattering, Analytical Ultracentrifugation and Constrained Modelling: A Comparison with Monomeric Human IgA1.
J.Mol.Biol., 338, 2004
|
|
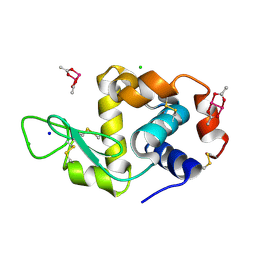
4OOO
 
 | |
4OT4
 
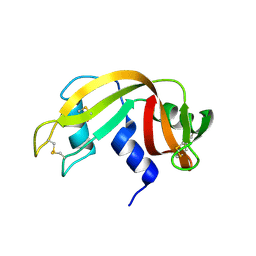 | |
4OVH
 
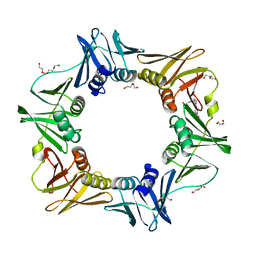 | | E. coli sliding clamp in complex with (R)-6-bromo-9-(2-(carboxymethylamino)-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-bromo-9-{2-[(carboxymethyl)amino]-2-oxoethyl}-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
6P1T
 
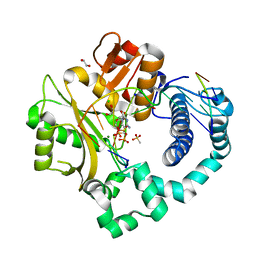 | | Pre-catalytic ternary complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and bound CMPCPP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
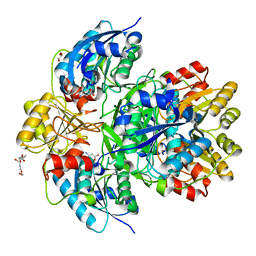
6PAE
 
 | |
6P1W
 
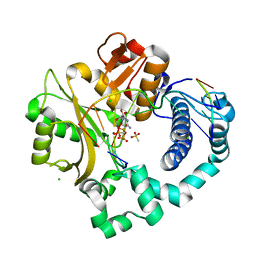 | | Pre-catalytic ternary complex of human DNA Polymerase Mu with 1-nt gapped substrate containing undamaged template dG and bound incoming CMPCPP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
4PPO
 
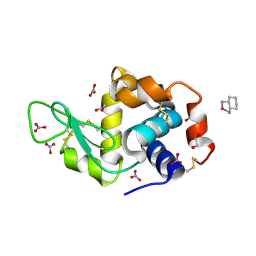 | | First Crystal Structure for an Oxaliplatin-Protein Complex | | Descriptor: | 1,2-ETHANEDIOL, CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), Lysozyme C, ... | | Authors: | Messori, L, Merlino, A. | | Deposit date: | 2014-02-27 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The X-ray structure of the complex formed in the reaction between oxaliplatin and lysozyme.
Chem.Commun.(Camb.), 50, 2014
|
|
4PFY
 
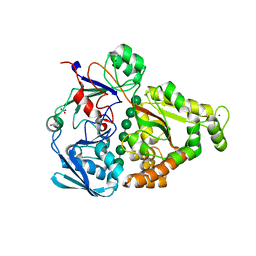 | | Crystal structure of mannohexaose bound oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (TM1223) from THERMOTOGA MARITIMA at 1.5 A resolution | | Descriptor: | ABC transporter substrate-binding protein, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Lu, X, Ghimire-Rijal, S, Cuneo, M.J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Duplication of Genes in an ATP-binding Cassette Transport System Increases Dynamic Range While Maintaining Ligand Specificity.
J.Biol.Chem., 289, 2014
|
|
6PCO
 
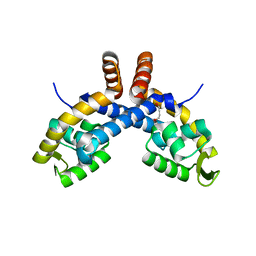 | | Mechanism for regulation of DNA binding of Bordetella bronchiseptica BpsR by 6-hydroxynicotinic acid | | Descriptor: | 1,4-BUTANEDIOL, MarR-family transcriptional regulator | | Authors: | Booth, W.T, Davis, R.R, Deora, R, Hollis, T. | | Deposit date: | 2019-06-17 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanism for regulation of DNA binding of BpsR, a Bordetella regulator of biofilm formation, by 6-hydroxynicotinic acid.
Plos One, 14, 2019
|
|
6PEI
 
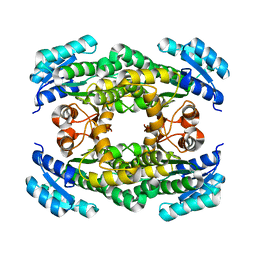 | | Structure of sorbitol dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Sorbitol dehydrogenase (L-iditol 2-dehydrogenase) | | Authors: | Bailey-Elkin, B.A, Kohlmeier, M.G, Oresnik, I.J, Mark, B.L. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the sorbitol dehydrogenase SmoS from Sinorhizobium meliloti 1021
Acta Crystallogr.,Sect.D, 77, 2021
|
|
6P1N
 
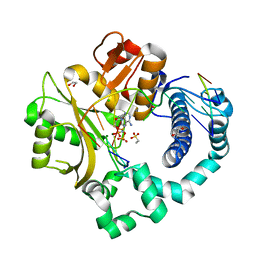 | | Pre-catalytic ternary complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and bound incoming dAMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
6P1S
 
 | | Post-catalytic nicked complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and newly incorporated AMP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
6PH6
 
 | |
6P1P
 
 | | Pre-catalytic ternary complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and bound incoming dCMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
6P1V
 
 | | Pre-catalytic ternary complex of human DNA Polymerase Mu with 1-nt gapped substrate containing undamaged template dG and bound incoming dCMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
