6M3D
 
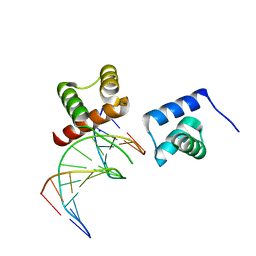 | | X-ray crystal structure of tandemly connected engrailed homeodomains (EHD) with R53A mutations and DNA complex | | Descriptor: | DNA (5'-D(*GP*GP*AP*TP*TP*AP*GP*GP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*TP*CP*CP*TP*AP*AP*TP*CP*C)-3'), SODIUM ION, ... | | Authors: | Sunami, T, Hirano, Y, Tamada, T, Kono, H. | | Deposit date: | 2020-03-03 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for designing an array of engrailed homeodomains.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6QMS
 
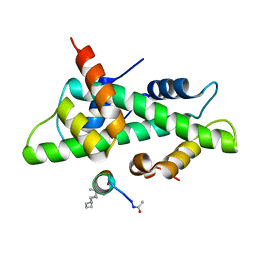 | | NF-YB/C Heterodimer in Complex with NF-YA-derived Peptide Stabilized with C11-Hydrocarbon Linker | | Descriptor: | Nuclear transcription factor Y subunit alpha, Nuclear transcription factor Y subunit beta, Nuclear transcription factor Y subunit gamma | | Authors: | Kiehstaller, S, Jeganathan, S, Pearce, N.M, Wendt, M, Grossmann, T.N, Hennig, S. | | Deposit date: | 2019-02-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Constrained Peptides with Fine-Tuned Flexibility Inhibit NF-Y Transcription Factor Assembly.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6SQK
 
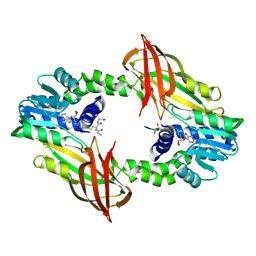 | |
6SQH
 
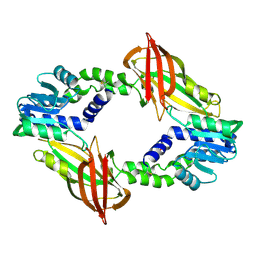 | |
6QDR
 
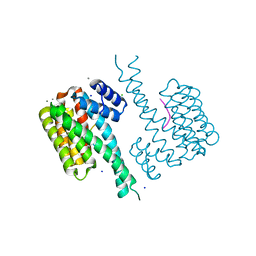 | | Crystal structure of 14-3-3sigma in complex with a PAK6 pT99 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Kaplan, A, Fournier, A.E, Ottman, C. | | Deposit date: | 2019-01-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.615 Å) | | Cite: | Polypharmacological Perturbation of the 14-3-3 Adaptor Protein Interactome Stimulates Neurite Outgrowth.
Cell Chem Biol, 27, 2020
|
|
6N0J
 
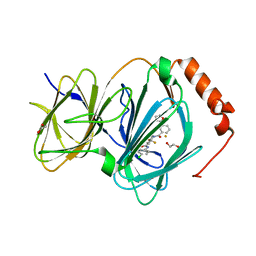 | | The complex of CCG-222740 bound to pirin | | Descriptor: | (3S)-N-(4-chlorophenyl)-5,5-difluoro-1-[3-(furan-2-yl)benzene-1-carbonyl]piperidine-3-carboxamide, 1,2-ETHANEDIOL, FE (III) ION, ... | | Authors: | Lisabeth, E.M, Jin, X, Neubig, R. | | Deposit date: | 2018-11-07 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of Pirin as a Molecular Target of the CCG-1423/CCG-203971 Series of Antifibrotic and Antimetastatic Compounds
ACS Pharmacol Transl Sci, 2, 2019
|
|
6N2G
 
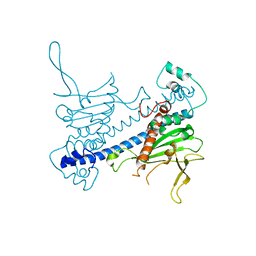 | | Crystal structure of Caenorhabditis elegans NAP1 | | Descriptor: | Nucleosome Assembly Protein | | Authors: | Bhattacharyya, S, DArcy, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Characterization of Caenorhabditis elegans Nucleosome Assembly Protein 1 Uncovers the Role of Acidic Tails in Histone Binding.
Biochemistry, 58, 2019
|
|
3D87
 
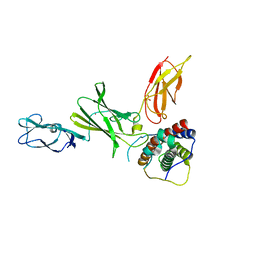 | | Crystal structure of Interleukin-23 | | Descriptor: | Interleukin-12 subunit p40, Interleukin-23 subunit p19, PHOSPHATE ION, ... | | Authors: | Beyer, B.M, Ingram, R, Ramanathan, L, Reichert, P, Le, H, Madison, V. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the pro-inflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody
J.Mol.Biol., 382, 2008
|
|
3D85
 
 | | Crystal structure of IL-23 in complex with neutralizing FAB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FAB of antibody 7G10, heavy chain, ... | | Authors: | Beyer, B.M, Ingram, R, Ramanathan, L, Reichert, P, Le, H, Madison, V. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the pro-inflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody
J.Mol.Biol., 382, 2008
|
|
6RSU
 
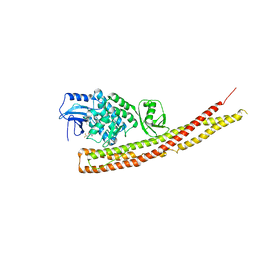 | | TBK1 in complex with Inhibitor compound 35 | | Descriptor: | 3,3,3-tris(fluoranyl)-1-[4-[(1~{R})-1-[2-[[(2~{S})-5-(5-propan-2-yloxypyrimidin-4-yl)-2,3-dihydro-1~{H}-benzimidazol-2-yl]amino]pyridin-4-yl]ethyl]piperazin-1-yl]propan-1-one, Serine/threonine-protein kinase TBK1 | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6N7H
 
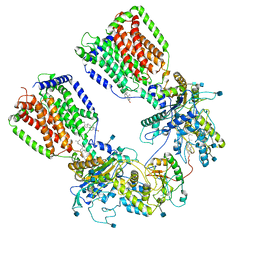 | | Cryo-EM structure of the 2:1 hPtch1-Shhp complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm.
Nat Commun, 10, 2019
|
|
4GIZ
 
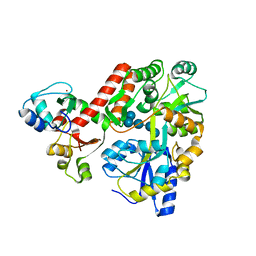 | | Crystal structure of full-length human papillomavirus oncoprotein E6 in complex with LXXLL peptide of ubiquitin ligase E6AP at 2.55 A resolution | | Descriptor: | Maltose-binding periplasmic protein, UBIQUITIN LIGASE EA6P: chimeric protein, Protein E6, ... | | Authors: | McEwen, A.G, Zanier, K, Charbonnier, S, Poussin, P, Cura, V, Vande Pol, S, Trave, G, Cavarelli, J. | | Deposit date: | 2012-08-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins.
Science, 339, 2013
|
|
6SIV
 
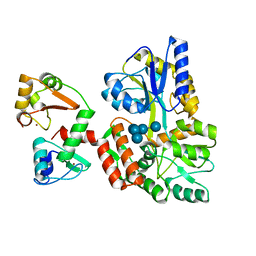 | | Structure of HPV16 E6 oncoprotein in complex with mutant IRF3 LxxLL motif | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | Authors: | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | Deposit date: | 2019-08-12 | | Release date: | 2019-08-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Deciphering the molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
6SKO
 
 | | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork - conformation 2 MCM CTD:ssDNA | | Descriptor: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | Authors: | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
6SMA
 
 | | Crystal structure of Human Neutrophil Elastase (HNE) in complex with the 3-Oxo-beta-Sultam inhibitor LMC249 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[1-[(4-bromophenyl)methyl]-1,2,3-triazol-4-yl]methylcarbamoyl]pentane-3-sulfonic acid, ... | | Authors: | Brito, J.A, Almeida, V.T, Carvalho, L.M, Moreira, R, Archer, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | 3-Oxo-beta-sultam as a Sulfonylating Chemotype for Inhibition of Serine Hydrolases and Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
6SXA
 
 | | XPF-ERCC1 Cryo-EM Structure, Apo-form | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Jones, M.L, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2019-09-25 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of the XPF-ERCC1 endonuclease reveal how DNA-junction engagement disrupts an auto-inhibited conformation.
Nat Commun, 11, 2020
|
|
6SG4
 
 | | Structure of CDK2/cyclin A M246Q, S247EN | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Salamina, M, Basle, A, Massa, B, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2019-08-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discriminative SKP2 Interactions with CDK-Cyclin Complexes Support a Cyclin A-Specific Role in p27KIP1 Degradation.
J.Mol.Biol., 433, 2021
|
|
6RVD
 
 | | Revised cryo-EM structure of the human 2:1 Ptch1-Shh complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | El Omari, K, Rudolf, A.F, Kowatsch, C, Malinauskas, T, Kinnebrew, M, Ansell, T.B, Bishop, B, Pardon, E, Schwab, R.A, Qian, M, Duman, R, Covey, D.F, Steyaert, J, Wagner, A, Sansom, M.S.P, Rohatgi, R, Siebold, C. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism.
Nat.Chem.Biol., 15, 2019
|
|
6SLX
 
 | | Fragment AZ-010 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, TAZpS89, ~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]propanamide | | Authors: | Ottmann, C, Wolter, M, Guillory, X, Genet, S, Somsen, B, Leysen, S, Castaldi, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.800045 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6SVK
 
 | | human Myeloid-derived growth factor (MYDGF) | | Descriptor: | MALONATE ION, Myeloid-derived growth factor | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2019-09-18 | | Release date: | 2019-11-27 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Crystal structure and receptor-interacting residues of MYDGF - a protein mediating ischemic tissue repair.
Nat Commun, 10, 2019
|
|
6SQ4
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor U2 | | Descriptor: | 9-(7-{[amino(iminio)methyl]amino}-5,6,7-trideoxy-beta-D-ribo-heptofuranosyl)-9H-purin-6-amine, IODIDE ION, Protein arginine N-methyltransferase 6 | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of mouse PRMT6 in complex with inhibitors
To be published
|
|
6SQ3
 
 | | Crystal structure of mouse PRMT6 in complex with inhibitor U1 | | Descriptor: | Protein arginine N-methyltransferase 6, [[2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethylamino]-azanyl-methylidene]azanium | | Authors: | Bonnefond, L, Cavarelli, J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of mouse PRMT6 in complex with inhibitors
To Be Published
|
|
6SQI
 
 | |
8CPD
 
 | | Cryo-EM structure of CRaf dimer with 14:3:3 | | Descriptor: | 14-3-3 protein zeta isoform X1, RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Dedden, D, Graedler, U, Schwarz, D, Thomsen, M, Leuthner, B, Schneider, E, Nitsche, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
8CHF
 
 | | cryo-EM Structure of Craf:14-3-3:Mek1 | | Descriptor: | 14-3-3 protein zeta isoform X1, 2-{4-[(1E)-1-(hydroxyimino)-2,3-dihydro-1H-inden-5-yl]-3-(pyridin-4-yl)-1H-pyrazol-1-yl}ethanol, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Dedden, D, Ulrich, G. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
