5UFG
 
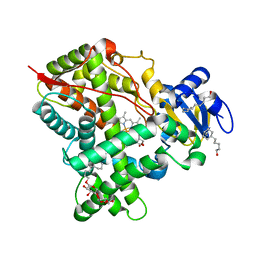 | | Crystal Structure of CYP2B6 (Y226H/K262R/I114V) in complex with myrtenyl bromide | | Descriptor: | (1S,5R)-2-(bromomethyl)-6,6-dimethylbicyclo[3.1.1]hept-2-ene, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Halpert, J.R. | | Deposit date: | 2017-01-04 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Halogen-pi Interactions in the Cytochrome P450 Active Site: Structural Insights into Human CYP2B6 Substrate Selectivity.
ACS Chem. Biol., 12, 2017
|
|
7SKX
 
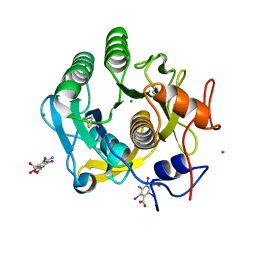 | | Ab initio structure of proteinase K from electron-counted MicroED data | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CALCIUM ION, Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Hattne, J, Gonen, T. | | Deposit date: | 2021-10-21 | | Release date: | 2022-06-08 | | Last modified: | 2022-06-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.5 Å) | | Cite: | Ab initio phasing macromolecular structures using electron-counted MicroED data.
Nat.Methods, 19, 2022
|
|
4PR6
 
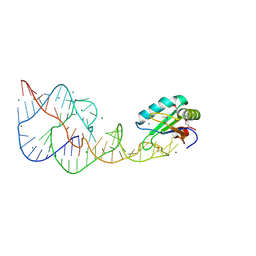 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | HDV RIBOZYME SELF-CLEAVED, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2014-11-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
6Q8Q
 
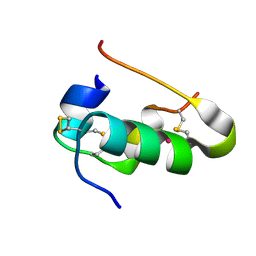 | |
4ZHF
 
 | | Siderocalin-mediated recognition and cellular uptake of actinides | | Descriptor: | CURIUM ION, GLYCEROL, N,N'-butane-1,4-diylbis[1-hydroxy-N-(3-{[(1-hydroxy-6-oxo-1,6-dihydropyridin-2-yl)carbonyl]amino}propyl)-6-oxo-1,6-dihydropyridine-2-carboxamide], ... | | Authors: | Allred, B.E, Rupert, P.B, Gauny, S.S, An, D.D, Ralston, C.Y, Sturzbecher-Hoehne, M, Strong, R.K, Abergel, R.J. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Siderocalin-mediated recognition, sensitization, and cellular uptake of actinides.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6Q8T
 
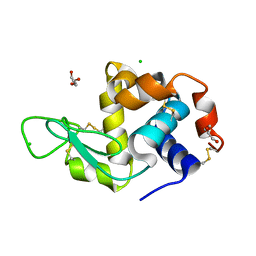 | | Cryo structure of HEWL at 81 kGy | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Lysozyme C | | Authors: | de la Mora, E, Coquelle, N, Bury, C.S, Rosenthal, M, Garman, E.F, Burghammer, M, Colletier, J.P, Weik, M. | | Deposit date: | 2018-12-16 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74008667 Å) | | Cite: | Radiation damage and dose limits in serial synchrotron crystallography at cryo- and room temperatures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4Z3M
 
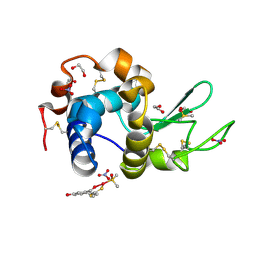 | | X-ray structure of the adduct formed in the reaction between lysozyme and a platinum(II) Complex with S,O Bidentate Ligands (9b) | | Descriptor: | 1,2-ETHANEDIOL, 3-[2-chloranyl-2-[dimethyl(oxidanyl)-{4}-sulfanyl]-4-ethylsulfanyl-1-oxa-3{3}-thia-2{4}-platinacyclohexa-3,5-dien-6-yl]phenol, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A. | | Deposit date: | 2015-03-31 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Platinum(II) Complexes with O,S Bidentate Ligands: Biophysical Characterization, Antiproliferative Activity, and Crystallographic Evidence of Protein Binding.
Inorg.Chem., 54, 2015
|
|
6QEY
 
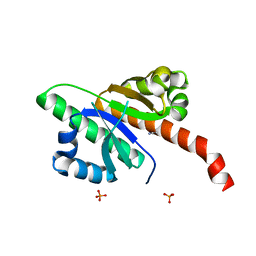 | | IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties | | Descriptor: | ACETONITRILE, Insulin-like growth factor 2 mRNA-binding protein 1, PHOSPHATE ION | | Authors: | Dagil, R, Ball, N.J, Ogrodowicz, R.W, Purkiss, A.G, Taylor, I.A, Ramos, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties.
Nucleic Acids Res., 47, 2019
|
|
4PHH
 
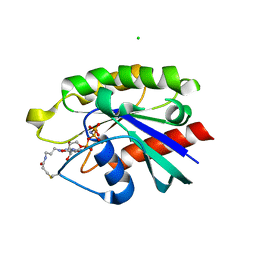 | | Crystal structure of Ypt7 covalently modified with GNP | | Descriptor: | 5'-O-[(R)-hydroxy{[(S)-hydroxy(phosphonoamino)phosphoryl]oxy}phosphoryl]-N-[3-(propanoylamino)propyl]guanosine, CHLORIDE ION, GTP-binding protein YPT7, ... | | Authors: | Wiegandt, D, Vieweg, S, Hofmann, F, Koch, D, Wu, Y, Itzen, A, Mueller, M.P, Goody, R.S. | | Deposit date: | 2014-05-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Locking GTPases covalently in their functional states.
Nat Commun, 6, 2015
|
|
4ZID
 
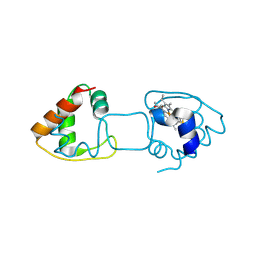 | | Dimeric Hydrogenobacter thermophilus cytochrome c552 obtained from Escherichia coli | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Hayashi, Y, Yamanaka, M, Nagao, S, Komori, H, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-04-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Domain swapping oligomerization of thermostable c-type cytochrome in E. coli cells
Sci Rep, 6, 2016
|
|
6RON
 
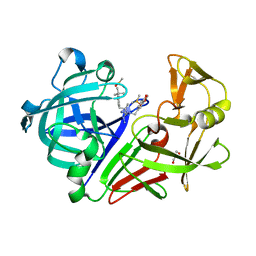 | |
5TUR
 
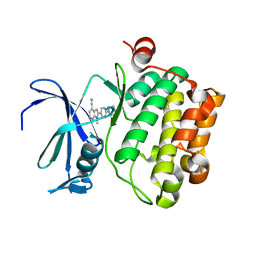 | | Pim-1 kinase in complex with a 7-azaindole | | Descriptor: | 1-methyl-2-[4-(piperazin-1-yl)phenyl]-1H-pyrrolo[2,3-b]pyridine-4-carbonitrile, Serine/threonine-protein kinase pim-1 | | Authors: | Mechin, I, Zhang, Y, Wang, R, Batchelor, J.D, Mclean, L. | | Deposit date: | 2016-11-07 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.948 Å) | | Cite: | Discovery of N-substituted 7-azaindoles as PIM1 kinase inhibitors - Part I.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6RFP
 
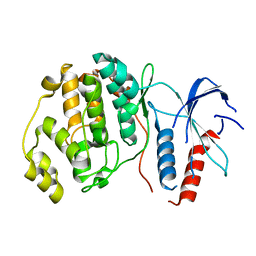 | | ERK2 MAP kinase with mutations at Helix-G | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Livnah, O, Eitan-Wexler, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The bacterial metalloprotease NleD selectively cleaves mitogen-activated protein kinases that have high flexibility in their activation loop.
J.Biol.Chem., 295, 2020
|
|
6FTZ
 
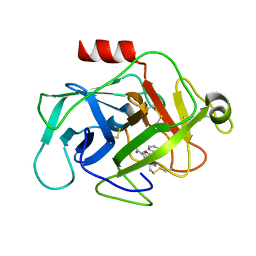 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 6 | | Descriptor: | Complement factor D, ~{N}4-[3-(aminomethyl)phenyl]-1~{H}-indole-2,4-dicarboxamide | | Authors: | Ostermann, N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
5U06
 
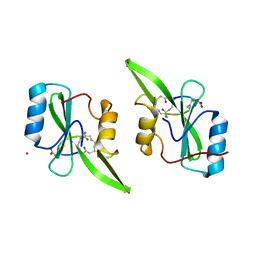 | | Grb7-SH2 with bicyclic peptide inhibitor containing a pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, POTASSIUM ION, bicyclic peptide inhibitor: LYS-PHE-GLU-GLY-CMF-ASP-ASN-GLU-CST | | Authors: | Watson, G.M, Wilce, J.A. | | Deposit date: | 2016-11-22 | | Release date: | 2017-11-15 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
6FYL
 
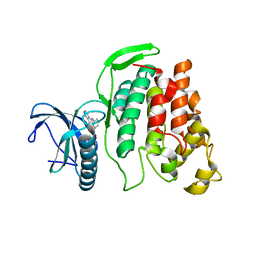 | | X-ray structure of CLK2-KD(136-496)/CX-4945 at 1.95A | | Descriptor: | 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, Dual specificity protein kinase CLK2 | | Authors: | Kallen, J. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray Structures and Feasibility Assessment of CLK2 Inhibitors for Phelan-McDermid Syndrome.
ChemMedChem, 13, 2018
|
|
6FUH
 
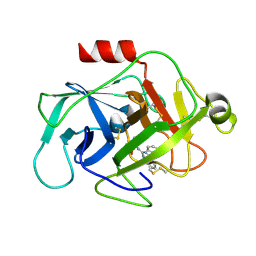 | | Complement factor D in complex with the inhibitor (4-((3-(aminomethyl)phenyl)amino)quinazolin-2-yl)-L-valine | | Descriptor: | (2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]-3-methyl-butanoic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
7RWE
 
 | | Crystal structure of CDK2 liganded with compound GPHR787 | | Descriptor: | 5-nitro-2-[(3-phenylpropyl)amino]benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
6RT1
 
 | | Native tetragonal lysozyme - home source data | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Pereira, P.J.B. | | Deposit date: | 2019-05-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.336 Å) | | Cite: | Protein crystals as a key for deciphering macromolecular crowding effects on biological reactions.
Phys Chem Chem Phys, 22, 2020
|
|
7S2V
 
 | |
4Z8Y
 
 | | Crystal structure of Rab GTPase Sec4p mutant - S29V | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein SEC4, ... | | Authors: | Rinadi, F.C, Collins, R.N. | | Deposit date: | 2015-04-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the molecular mechanism of the Rab GTPase Sec4p activation.
Bmc Struct.Biol., 15, 2015
|
|
6G01
 
 | |
6FMA
 
 | | Crystal structure of ERK2 in complex with an adenosine derivative | | Descriptor: | 3-[6-azanyl-9-[(2~{R},3~{R},4~{S},5~{R})-5-(azidomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]sulfanylpropanoic acid, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 1, ... | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2018-01-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.667 Å) | | Cite: | None
To be published
|
|
5T2R
 
 | |
6FMO
 
 | |
