1GRG
 
 | |
1GRE
 
 | |
6Z9W
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLGWVFAQV | | Descriptor: | Beta-2-microglobulin, LEU-LEU-GLY-TRP-VAL-PHE-ALA-GLN-VAL, MHC class I antigen | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
6Z9V
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting IIGWMWIPV | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
6Z9X
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLS (t-butyl)Y FGTPT | | Descriptor: | Beta-2-microglobulin, LEU-LEU-SER-TUR-PHE-GLY-THR-PRO-THR, MHC class I antigen, ... | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
1HKC
 
 | |
1HRH
 
 | |
6Z01
 
 | | DNA Topoisomerase | | Descriptor: | CHLORIDE ION, DNA topoisomerase I | | Authors: | Takahashi, T.S, Gadelle, D, Forterre, P, Mayer, C, Petrella, S. | | Deposit date: | 2020-05-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states.
Nat Commun, 13, 2022
|
|
6Z03
 
 | | DNA Topoisomerase | | Descriptor: | DNA topoisomerase I | | Authors: | Takahashi, T.S, Gadelle, D, Forterre, P, Mayer, C, Petrella, S. | | Deposit date: | 2020-05-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states.
Nat Commun, 13, 2022
|
|
6YW8
 
 | |
6ZSC
 
 | | Human mitochondrial ribosome in complex with E-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
6ZM6
 
 | | Human mitochondrial ribosome in complex with mRNA, A/A tRNA and P/P tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Andrell, J, Amunts, A. | | Deposit date: | 2020-07-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Mechanism of membrane-tethered mitochondrial protein synthesis.
Science, 371, 2021
|
|
1HM8
 
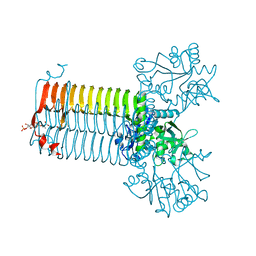 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE, GLMU, BOUND TO ACETYL COENZYME A | | Descriptor: | ACETYL COENZYME *A, CALCIUM ION, UDP-N-ACETYLGLUCOSAMINE-1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-05 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
6ZYB
 
 | |
7A5J
 
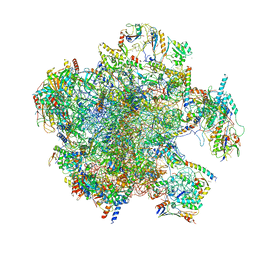 | | Structure of the split human mitoribosomal large subunit with P-and E-site mt-tRNAs | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Desai, N, Yang, H, Chandrasekaran, V, Kazi, R, Minczuk, M, Ramakrishnan, V. | | Deposit date: | 2020-08-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elongational stalling activates mitoribosome-associated quality control.
Science, 370, 2020
|
|
1HW6
 
 | |
1HRK
 
 | | CRYSTAL STRUCTURE OF HUMAN FERROCHELATASE | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, FERROCHELATASE | | Authors: | Wu, C.K, Dailey, H.A, Rose, J.P, Burden, A, Sellers, V.M, Wang, B.-C. | | Deposit date: | 2000-12-21 | | Release date: | 2001-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
6ZTJ
 
 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
1HM0
 
 | | CRYSTAL STRUCTURE OF S.PNEUMONIAE N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE, GLMU | | Descriptor: | CALCIUM ION, N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Authors: | Sulzenbacher, G, Gal, L, Peneff, C, Fassy, F, Bourne, Y. | | Deposit date: | 2000-12-04 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J.Biol.Chem., 276, 2001
|
|
6ZTP
 
 | | E. coli 70S-RNAP expressome complex in uncoupled state 6 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
1HQT
 
 | | THE CRYSTAL STRUCTURE OF AN ALDEHYDE REDUCTASE Y50F MUTANT-NADP COMPLEX AND ITS IMPLICATIONS FOR SUBSTRATE BINDING | | Descriptor: | ALDEHYDE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Hyndman, D, Green, N.C, Li, L, Korithoski, B, Jia, Z, Flynn, T.G. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of an Aldehyde Reductase Y50F Mutant-NADP Complex and its Implications for Substrate Binding
Chem.Biol.Interact., 132, 2001
|
|
1HUI
 
 | | INSULIN MUTANT (B1, B10, B16, B27)GLU, DES-B30, NMR, 25 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Olsen, H.B, Ludvigsen, S, Kaarsholm, N.C. | | Deposit date: | 1996-03-29 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an engineered insulin monomer at neutral pH.
Biochemistry, 35, 1996
|
|
3SX4
 
 | | Crystal structure of human dpp-iv in complex with sa-(+)-3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)- 2-methyl-5h-pyrrolo[3,4-b]pyridin-7(6h)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)-2-methyl-5,6-dihydro-7H-pyrrolo[3,4-b]pyridin-7-one, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-14 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 7-Oxopyrrolopyridine-derived DPP4 inhibitors-mitigation of CYP and hERG liabilities via introduction of polar functionalities in the active site.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SWW
 
 | | Crystal structure of human dpp-iv in complex with sa-(+)-3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)- 2-methyl-5h-pyrrolo[3,4-b]pyridin-7(6h)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyethyl)-2-methyl-5,6-dihydro-7H-pyrrolo[3,4-b]pyridin-7-one, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-14 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 7-Oxopyrrolopyridine-derived DPP4 inhibitors-mitigation of CYP and hERG liabilities via introduction of polar functionalities in the active site.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3VHK
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a back pocket binder | | Descriptor: | 1,2-ETHANEDIOL, Vascular endothelial growth factor receptor 2, {3-[(5-methyl-2-phenyl-1,3-oxazol-4-yl)methoxy]phenyl}methanol | | Authors: | Iwata, H, Oki, H, Okada, K, Takagi, T, Tawada, M, Miyazaki, Y, Imamura, S, Hori, A, Hixon, M.S, Kimura, H, Miki, H. | | Deposit date: | 2011-08-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A Back-to-Front Fragment-Based Drug Design Search Strategy Targeting the DFG-Out Pocket of Protein Tyrosine Kinases.
ACS MED.CHEM.LETT., 3, 2012
|
|
