7T1I
 
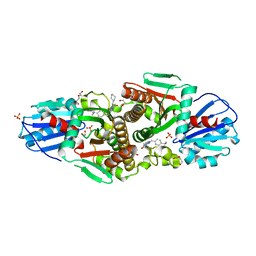 | | Crystal structure of CAB1 Pantothenate Kinase from Saccharomyces cerevisiae in complex with compound YU385597 | | Descriptor: | (8S)-2-{[(4-tert-butylphenyl)methyl]amino}-5-[(piperidin-1-yl)methyl][1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gihaz, S, Ben Mamoun, C. | | Deposit date: | 2021-12-02 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure and chemical screening reveal pantothenate kinase as a new target for antifungal development.
Structure, 30, 2022
|
|
7T1G
 
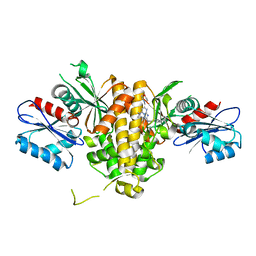 | | Crystal structure of CAB1 Pantothenate Kinase from Saccharomyces cerevisiae in complex with compound YU385595 | | Descriptor: | (8S)-N~2~-[(4-tert-butylphenyl)methyl]-N~7~,N~7~-dimethyl-5-[(morpholin-4-yl)methyl][1,2,4]triazolo[1,5-a]pyrimidine-2,7-diamine, Pantothenate kinase CAB1 | | Authors: | Gihaz, S, Ben Mamoun, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structure and chemical screening reveal pantothenate kinase as a new target for antifungal development.
Structure, 30, 2022
|
|
7MF4
 
 | |
5FXT
 
 | |
1TZM
 
 | | Crystal structure of ACC deaminase complexed with substrate analog b-chloro-D-alanine | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, 3-chloro-D-alanine, AMINO-ACRYLATE, ... | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-10 | | Release date: | 2004-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
7UL4
 
 | | CryoEM Structure of Inactive MOR Bound to Alvimopan and Mb6 | | Descriptor: | Megabody 6, Mu-type opioid receptor, N-[(2S)-2-{[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]methyl}-3-phenylpropanoyl]glycine | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7FV8
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z964297186 | | Descriptor: | 4-(3-chlorobenzoyl)-N-[3-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-3-oxopropyl]-1,4-diazepane-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6HIV
 
 | | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the complete mitoribosome | | Descriptor: | 12S rRNA, 50S ribosomal protein L13, putative, ... | | Authors: | Ramrath, D.J.F, Niemann, M, Leibundgut, M, Bieri, P, Prange, C, Horn, E.K, Leitner, A, Boerhringer, D, Schneider, A, Ban, N. | | Deposit date: | 2018-08-31 | | Release date: | 2018-10-03 | | Last modified: | 2018-11-07 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes.
Science, 362, 2018
|
|
1TFU
 
 | |
1STM
 
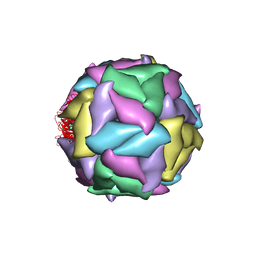 | | SATELLITE PANICUM MOSAIC VIRUS | | Descriptor: | SATELLITE PANICUM MOSAIC VIRUS | | Authors: | Ban, N, McPherson, A. | | Deposit date: | 1995-07-12 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of satellite panicum mosaic virus at 1.9 A resolution.
Nat.Struct.Biol., 2, 1995
|
|
3G9X
 
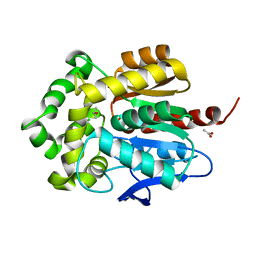 | | Structure of haloalkane dehalogenase DhaA14 mutant I135F from Rhodococcus rhodochrous | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Gavira, J.A, Stsiapanava, A, Kuty, M, Lapkouski, M, Dohnalek, J, Kuta Smatanova, I. | | Deposit date: | 2009-02-15 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3FWH
 
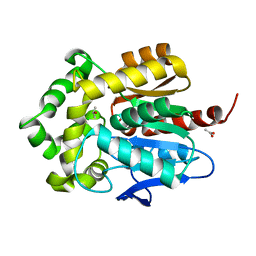 | | Structure of haloalkane dehalogenase mutant Dha15 (I135F/C176Y) from Rhodococcus rhodochrous | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Gavira, J.A, Stsiapanava, A, Kuty, M, Dohnalek, J, Lapkouski, M, Kuta Smatanova, I. | | Deposit date: | 2009-01-18 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7GOQ
 
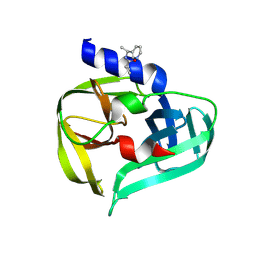 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z220996120 | | Descriptor: | DIMETHYL SULFOXIDE, N-[(1S)-1-(pyridin-3-yl)ethyl]cyclopropanecarboxamide, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GOX
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z33452106 | | Descriptor: | DIMETHYL SULFOXIDE, Protease 3C, ~{N}-(4-methyl-2-oxidanyl-phenyl)propanamide | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GP6
 
 | | PanDDA analysis group deposition -- Crystal Structure of Enterovirus D68 3C Protease in complex with Z53116498 | | Descriptor: | 3-(2-methyl-1H-benzimidazol-1-yl)propanamide, DIMETHYL SULFOXIDE, Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Thompson, W, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1N2O
 
 | |
1OD6
 
 | | The Crystal Structure of Phosphopantetheine adenylyltransferase from Thermus Thermophilus in complex with 4'-phosphopantetheine | | Descriptor: | 4'-PHOSPHOPANTETHEINE, PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE, SULFATE ION | | Authors: | Takahashi, H, Inagaki, E, Miyano, M, Tahirov, T.H. | | Deposit date: | 2003-02-13 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Implications for the Thermal Stability of Phosphopantetheine Adenylyltransferase from Thermus Thermophilus.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1C49
 
 | | STRUCTURAL AND FUNCTIONAL DIFFERENCES OF TWO TOXINS FROM THE SCORPION PANDINUS IMPERATOR | | Descriptor: | TOXIN K-BETA | | Authors: | Klenk, K.C, Tenenholz, T.C, Matteson, D.R, Rogowski, R.S, Blaustein, M.P, Weber, D.J. | | Deposit date: | 1999-08-17 | | Release date: | 2000-03-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural and functional differences of two toxins from the scorpion Pandinus imperator.
Proteins, 38, 2000
|
|
5OML
 
 | | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules to investigate the water envelope | | Descriptor: | (3~{R})-3-[[1-(2-hydroxyethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridin-3-yl]carbonylamino]-3-phenyl-propanoic acid, BETA-MERCAPTOETHANOL, Peroxin 14, ... | | Authors: | Ratkova, E.L, Dawidowski, M, Napolitano, V, Dubin, G, Fino, R, Popowicz, G, Sattler, M, Tetko, I.V. | | Deposit date: | 2017-08-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma Brucei PEX14 N-terminal domain in complex with small molecules to investigate the water envelope
To Be Published
|
|
5OW2
 
 | | Japanese encephalitis virus capsid protein | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, capsid protein | | Authors: | Poonsiri, T, Wright, G.S.A, Antonyuk, S.V. | | Deposit date: | 2017-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of the Japanese Encephalitis Virus Capsid Protein.
Viruses, 11, 2019
|
|
7YXO
 
 | | Crystal structure of WT AncGR2-LBD bound to dexamethasone and SHP coregulator fragment | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ancestral Glucocorticoid Receptor2, DEXAMETHASONE, ... | | Authors: | Jimenez-Panizo, A, Estebanez-Perpina, E, Fuentes-Prior, P. | | Deposit date: | 2022-02-16 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The multivalency of the glucocorticoid receptor ligand-binding domain explains its manifold physiological activities.
Nucleic Acids Res., 50, 2022
|
|
4MQ6
 
 | |
5O19
 
 | |
4HZG
 
 | | Structure of haloalkane dehalogenase DhaA from Rhodococcus rhodochrous | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Stsiapanava, A, Weiss, M.S, Mesters, J.R, Kuta Smatanova, I. | | Deposit date: | 2012-11-15 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5QOS
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1187701032 | | Descriptor: | (3S)-N-(pyrimidin-2-yl)azepan-3-amine, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
