1XYP
 
 | |
2GD2
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
4RFN
 
 | | Crystal structure of ADCC-potent Rhesus macaque ANTIBODY JR4 in complex with HIV-1 CLADE A/E GP120 and M48 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB HEAVY CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, FAB LIGHT CHAIN OF ADCC ANTI-HIV-1 ANTIBODY JR4, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
1WJD
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, 38 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
3R12
 
 | |
3IVI
 
 | | Design and Synthesis of Potent BACE-1 Inhibitors with Cellular Activity: Structure-Activity Relationship of P1 Substituents | | Descriptor: | Beta-secretase 1, GLYCEROL, N-[(1S,2R)-3-{[(5S)-5-(3-tert-butylphenyl)-4,5,6,7-tetrahydro-1H-indazol-5-yl]amino}-1-(3,5-difluorobenzyl)-2-hydroxypropyl]acetamide, ... | | Authors: | Pan, H. | | Deposit date: | 2009-09-01 | | Release date: | 2010-01-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of cell potent BACE-1 inhibitors: structure-activity relationship of P1' substituents.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3A99
 
 | | Structure of PIM-1 kinase crystallized in the presence of P27KIP1 Carboxy-terminal peptide | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Morishita, D, Takami, M, Yoshikawa, S, Katayama, R, Sato, S, Kukimoto-Niino, M, Umehara, T, Shirouzu, M, Sekimizu, K, Yokoyama, S, Fujita, N. | | Deposit date: | 2009-10-22 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cell-permeable carboxyl-terminal p27(Kip1) peptide exhibits anti-tumor activity by inhibiting Pim-1 kinase
J.Biol.Chem., 286, 2011
|
|
3IE5
 
 | | Crystal structure of Hyp-1 protein from Hypericum perforatum (St John's wort) involved in hypericin biosynthesis | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Michalska, K, Fernandes, H, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Crystal structure of Hyp-1, a St. John's wort protein implicated in the biosynthesis of hypericin
J.Struct.Biol., 169, 2010
|
|
1TVY
 
 | | beta-1,4-galactosyltransferase mutant Met344His (M344H-Gal-T1) complex with UDP-galactose and manganese | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-1,4-galactosyltransferase 1, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Ramakrishnan, B, Boeggeman, E, Qasba, P.K. | | Deposit date: | 2004-06-30 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effect of the Met344His mutation on the conformational dynamics of bovine beta-1,4-galactosyltransferase: crystal structure of the Met344His mutant in complex with chitobiose
Biochemistry, 43, 2004
|
|
6CDI
 
 | | Cryo-EM structure at 3.6 A resolution of vaccine-elicited antibody vFP16.02 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein 120, ... | | Authors: | Acharya, P, Xu, K, Liu, K, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1.
Nat. Med., 24, 2018
|
|
1U3C
 
 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1A30
 
 | | HIV-1 PROTEASE COMPLEXED WITH A TRIPEPTIDE INHIBITOR | | Descriptor: | HIV-1 PROTEASE, TRIPEPTIDE GLU-ASP-LEU | | Authors: | Louis, J.M, Dyda, F, Nashed, N.T, Kimmel, A.R, Davies, D.R. | | Deposit date: | 1998-01-27 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease.
Biochemistry, 37, 1998
|
|
4JCO
 
 | |
4JDE
 
 | | Crystal structure of PUD-1/PUD-2 heterodimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein F15E11.1, ... | | Authors: | Luo, S, Ye, K. | | Deposit date: | 2013-02-25 | | Release date: | 2013-11-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of PUD-1 and PUD-2, two proteins up-regulated in a long-lived daf-2 mutant.
Plos One, 8, 2013
|
|
3H5U
 
 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor 1 | | Descriptor: | N-({3-[(5S)-5-tert-butyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-1,2-benzisothiazol-7-yl}methyl)methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1XQZ
 
 | | Crystal Structure of hPim-1 kinase at 2.1 A resolution | | Descriptor: | Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Qian, K.C, Wang, L, Hickey, E.R, Studts, J, Barringer, K, Peng, C, Kronkaitis, A, Li, J, White, A, Mische, S, Farmer, B. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Constitutive Activity and a Unique Nucleotide Binding Mode of Human Pim-1 Kinase.
J.Biol.Chem., 280, 2005
|
|
4JB9
 
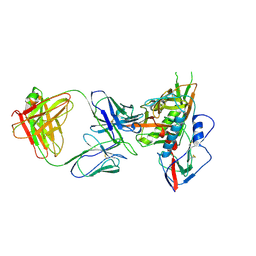 | | Crystal structure of antibody VRC06 in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, antibody VRC06 heavy chain, antibody VRC06 light chain, ... | | Authors: | Kwon, Y.D, Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-19 | | Release date: | 2013-05-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization.
Science, 340, 2013
|
|
2Z79
 
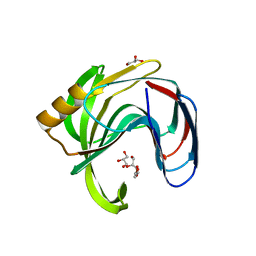 | | High resolution crystal structure of a glycoside hydrolase family 11 xylanase of Bacillus subtilis | | Descriptor: | Endo-1,4-beta-xylanase A, GLYCEROL | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
3AKI
 
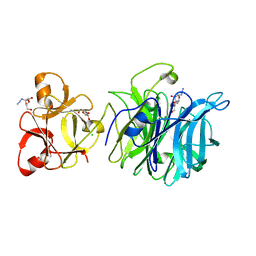 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-L-arabinofuranosyl azido | | Descriptor: | (2R,3R,4R,5S)-2-azido-5-(hydroxymethyl)oxolane-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
4OOZ
 
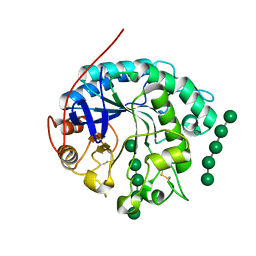 | | Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose | | Descriptor: | Beta-1,4-mannanase, beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, ... | | Authors: | Kim, M.-K, An, Y.J, Jeong, C.-S, Cha, S.-S. | | Deposit date: | 2014-02-04 | | Release date: | 2014-08-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based investigation into the functional roles of the extended loop and substrate-recognition sites in an endo-beta-1,4-d-mannanase from the Antarctic springtail, Cryptopygus antarcticus.
Proteins, 82, 2014
|
|
3AKF
 
 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3MVU
 
 | |
4EMZ
 
 | |
1ENX
 
 | |
1BV9
 
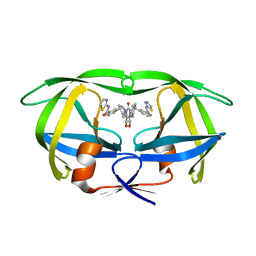 | | HIV-1 PROTEASE (I84V) COMPLEXED WITH XV638 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-09-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
