3P7A
 
 | | p38 inhibitor-bound | | Descriptor: | 1-[5-tert-butyl-2-(1,1-dioxidothiomorpholin-4-yl)thiophen-3-yl]-3-naphthalen-1-ylurea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Moffett, K.K, Namboodiri, H. | | Deposit date: | 2010-10-12 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a novel class of non-ATP site DFG-out state p38 inhibitors utilizing computationally assisted virtual fragment-based drug design (vFBDD).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1MAA
 
 | | MOUSE ACETYLCHOLINESTERASE CATALYTIC DOMAIN, GLYCOSYLATED PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Taylor, P, Bougis, P.E, Marchot, P. | | Deposit date: | 1998-11-04 | | Release date: | 1999-04-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of mouse acetylcholinesterase. A peripheral site-occluding loop in a tetrameric assembly.
J.Biol.Chem., 274, 1999
|
|
3OKN
 
 | | Crystal structure of S25-39 in complex with Kdo(2.4)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
4LVA
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[4-(pyrrolidin-1-yl)piperidin-1-yl]sulfonyl}benzyl)-2H-pyrido[4,3-e][1,2,4]thiadiazin-3-amine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8BN3
 
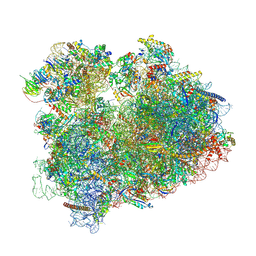 | | Yeast 80S, ES7s delta, eIF5A, Stm1 containing | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Dimitrova-Paternoga, L, Paternoga, H, Wilson, D.N. | | Deposit date: | 2022-11-12 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Evolving precision: rRNA expansion segment 7S modulates translation velocity and accuracy in eukaryal ribosomes.
Nucleic Acids Res., 52, 2024
|
|
2W1X
 
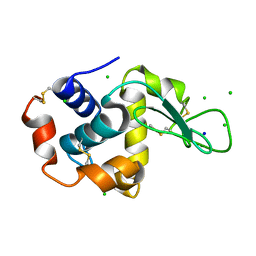 | | The interdependence of wavelength, redundancy and dose in sulfur SAD experiments: 1.284 A wavelength 360 images data | | Descriptor: | CHLORIDE ION, LYSOZYME C, SODIUM ION | | Authors: | Cianci, M, Helliwell, J.R, Suzuki, A. | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Interdependence of Wavelength, Redundancy and Dose in Sulfur Sad Experiments.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5PAM
 
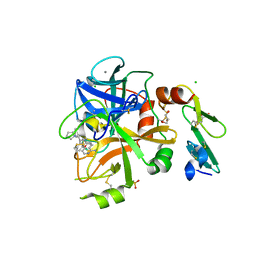 | | Crystal Structure of Factor VIIa in complex with 2-[(1-aminoisoquinolin-6-yl)amino]-2-(5-ethoxy-2-fluorophenyl)-1-(2-phenylpyrrolidin-1-yl)ethanone | | Descriptor: | (2~{R})-2-[(1-azanylisoquinolin-6-yl)amino]-2-(5-ethoxy-2-fluoranyl-phenyl)-1-[(2~{R})-2-phenylpyrrolidin-1-yl]ethanone, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-10 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
3WV6
 
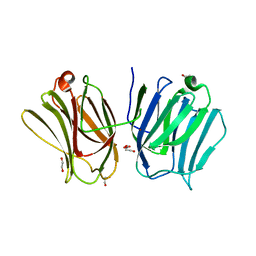 | |
3WVS
 
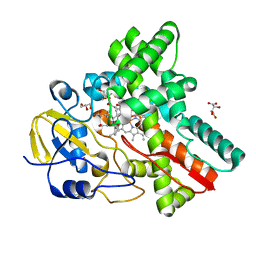 | | Crystal Structure of Cytochrome P450revI | | Descriptor: | (2E,4S,5S,6E,8E)-10-{(2R,3S,6S,8R,9S)-9-butyl-8-[(1E,3E)-4-carboxy-3-methylbuta-1,3-dien-1-yl]-3-methyl-1,7-dioxaspiro[5.5]undec-2-yl}-5-hydroxy-4,8-dimethyldeca-2,6,8-trienoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Nagano, S, Takahashi, S, Osada, H, Shiro, Y. | | Deposit date: | 2014-06-06 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses of cytochrome P450revI involved in reveromycin A biosynthesis and evaluation of the biological activity of its substrate, reveromycin T.
J.Biol.Chem., 289, 2014
|
|
9I0F
 
 | | Revisited AvNifEN crystal structure | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FeFe cofactor, ... | | Authors: | Paya Tormo, L, Nguyen, T.Q, Fyfe, C, Basbous, H, Dobrzynska, K, Echavarri-Erasun, C, Martin, L, Caserta, G, Legrand, P, Thorn, A, Amara, P, Schoehn, G, Cherrier, M.V, Rubio, L.M, Nicolet, Y. | | Deposit date: | 2025-01-15 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamics driving the precursor in NifEN scaffold during nitrogenase FeMo-cofactor assembly.
Nat.Chem.Biol., 2025
|
|
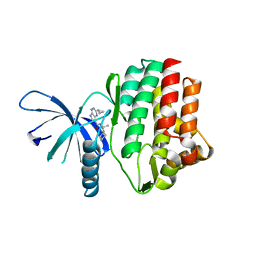
4HVI
 
 | |
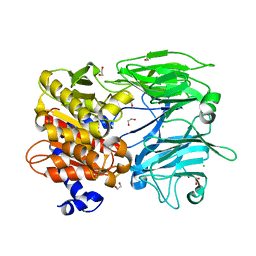
4HVT
 
 | |
3H6V
 
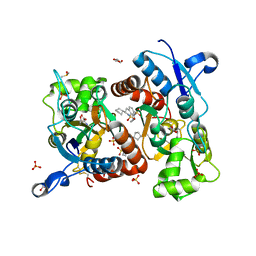 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5206 at 2.10 A resolution | | Descriptor: | (3R)-3-cyclopentyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
2IWI
 
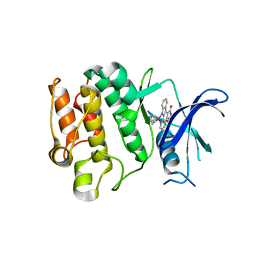 | | CRYSTAL STRUCTURE OF THE HUMAN PIM2 IN COMPLEX WITH A RUTHENIUM ORGANOMETALLIC LIGAND RU1 | | Descriptor: | RUTHENIUM-PYRIDOCARBAZOLE-1, SERINE/THREONINE-PROTEIN KINASE PIM-2 | | Authors: | Russo, S, Debreczeni, J.E, Amos, A, Bullock, A.N, Fedorov, O, Niesen, F, Sobott, F, Turnbull, A, Pike, A.C.W, Ugochukwu, E, Papagrigoriou, E, Bunkoczi, G, Gorrec, F, Edwards, A, Arrowsmith, C, Weigelt, J, Sundstrom, M, von Delft, F, Knapp, S. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the PIM2 kinase in complex with an organoruthenium inhibitor.
PLoS ONE, 4, 2009
|
|
1ZY6
 
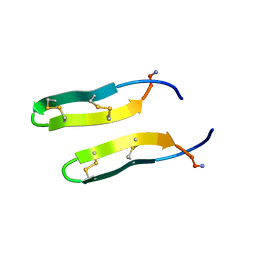 | | Membrane-bound dimer structure of Protegrin-1 (PG-1), a beta-Hairpin Antimicrobial Peptide in Lipid Bilayers from Rotational-Echo Double-Resonance Solid-State NMR | | Descriptor: | Protegrin 1 | | Authors: | Wu, X, Mani, R, Tang, M, Buffy, J.J, Waring, A.J, Sherman, M.A, Hong, M. | | Deposit date: | 2005-06-09 | | Release date: | 2006-06-13 | | Last modified: | 2024-11-13 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-Bound Dimer Structure of a beta-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR.
Biochemistry, 45, 2006
|
|
2J0M
 
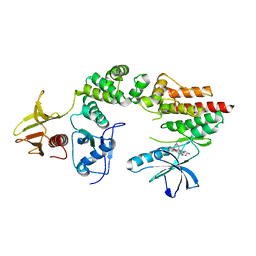 | | Crystal structure a two-chain complex between the FERM and kinase domains of focal adhesion kinase. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase.
Cell(Cambridge,Mass.), 129, 2007
|
|
2DQU
 
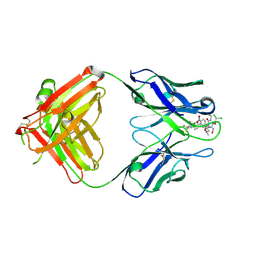 | | Crystal form II: high resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
1IVP
 
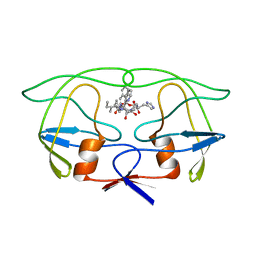 | | THE CRYSTALLOGRAPHIC STRUCTURE OF THE PROTEASE FROM HUMAN IMMUNODEFICIENCY VIRUS TYPE 2 WITH TWO SYNTHETIC PEPTIDIC TRANSITION STATE ANALOG INHIBITORS | | Descriptor: | 4-[(2R)-3-{[(1S,2S,3R,4S)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methyl-4-({(1S,2R)-2-methyl-1-[(pyridin-2-ylmethyl)carba moyl]butyl}carbamoyl)hexyl]amino}-2-{[(naphthalen-1-yloxy)acetyl]amino}-3-oxopropyl]-1H-imidazol-3-ium, HIV-2 PROTEASE | | Authors: | Mulichak, A.M, Watenpaugh, K.D. | | Deposit date: | 1993-03-18 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystallographic structure of the protease from human immunodeficiency virus type 2 with two synthetic peptidic transition state analog inhibitors.
J.Biol.Chem., 268, 1993
|
|
2J0J
 
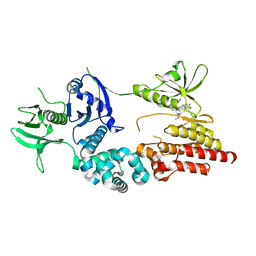 | | Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase
Cell(Cambridge,Mass.), 129, 2007
|
|
6J71
 
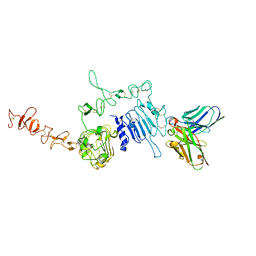 | | HuA21-scFv in complex with the extracellular domain(ECD) of HER2 | | Descriptor: | Receptor tyrosine-protein kinase erbB-2, anti-HER2 humanized antibody HuA21, beta-D-mannopyranose, ... | | Authors: | Wang, Z, Guo, G, Cheng, B, Zhu, Z, Niu, L, Zhang, H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-02-27 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Structural insight into a matured humanized monoclonal antibody HuA21 against HER2-overexpressing cancer cells.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4IA2
 
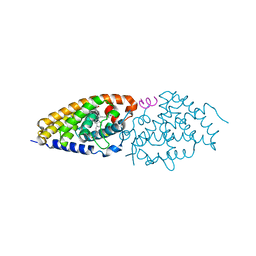 | | Diastereotopic and Deuterium Effects in Gemini | | Descriptor: | 21-NOR-9,10-SECOCHOLESTA-5,7,10(19)-TRIENE-1,3,25-TRIOL, 20-(4-HYDROXY-4-METHYLPENTYL)-, (1A,3B,5Z,7E), ... | | Authors: | Maehr, H, Rochel, N, Suh, N, Uskokovic, M. | | Deposit date: | 2012-12-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Diastereotopic and deuterium effects in gemini.
J.Med.Chem., 56, 2013
|
|
4PY8
 
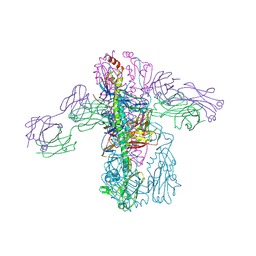 | | Crystal structure of Fab 3.1 in complex with the 1918 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | Dreyfus, C. | | Deposit date: | 2014-03-26 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Alternative Recognition of the Conserved Stem Epitope in Influenza A Virus Hemagglutinin by a VH3-30-Encoded Heterosubtypic Antibody.
J.Virol., 88, 2014
|
|
1R10
 
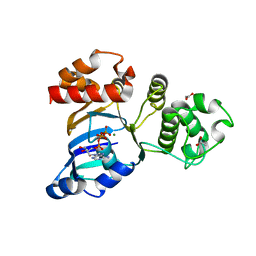 | | Cystic fibrosis transmembrane conductance regulator (CFTR) nucleotide-binding domain one (NBD1) with ATP, I4122 space group | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
2J0K
 
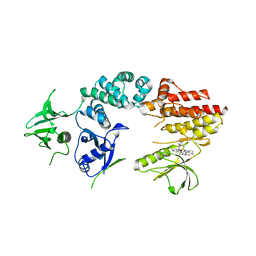 | | Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase.
Cell(Cambridge,Mass.), 129, 2007
|
|
6J4W
 
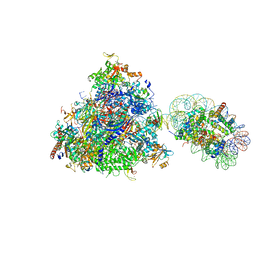 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
