2HVJ
 
 | | Crystal structure of KcsA-Fab-TBA complex in low K+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel.
J.Mol.Biol., 366, 2007
|
|
2HVK
 
 | | crystal structure of the KcsA-Fab-TBA complex in high K+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, Antibody Fab heavy chain, Antibody Fab light chain, ... | | Authors: | Zhou, Y. | | Deposit date: | 2006-07-28 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel.
J.Mol.Biol., 366, 2007
|
|
2DWD
 
 | | crystal structure of KcsA-FAB-TBA complex in Tl+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
2DWE
 
 | | Crystal structure of KcsA-FAB-TBA complex in Rb+ | | Descriptor: | (2S)-3-HYDROXY-2-(NONANOYLOXY)PROPYL LAURATE, ANTIBODY FAB HEAVY CHAIN, ANTIBODY FAB LIGHT CHAIN, ... | | Authors: | Yohannan, S, Zhou, Y. | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic Study of the Tetrabutylammonium Block to the KcsA K(+) Channel
J.Mol.Biol., 366, 2007
|
|
1L2I
 
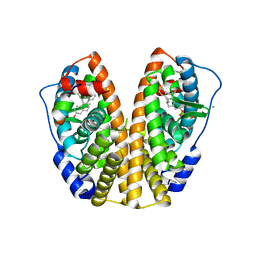 | | Human Estrogen Receptor alpha Ligand-binding Domain in Complex with (R,R)-5,11-cis-diethyl-5,6,11,12-tetrahydrochrysene-2,8-diol and a Glucocorticoid Receptor Interacting Protein 1 NR box II Peptide | | Descriptor: | (R,R)-5,11-CIS-DIETHYL-5,6,11,12-TETRAHYDROCHRYSENE-2,8-DIOL, CHLORIDE ION, ESTROGEN RECEPTOR, ... | | Authors: | Shiau, A.K, Barstad, D, Radek, J.T, Meyers, M.J, Nettles, K.W, Katzenellenbogen, B.S, Katzenellenbogen, J.A, Agard, D.A, Greene, G.L. | | Deposit date: | 2002-02-21 | | Release date: | 2002-05-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism.
Nat.Struct.Biol., 9, 2002
|
|
1DFO
 
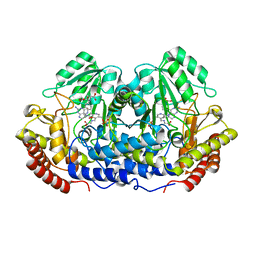 | | CRYSTAL STRUCTURE AT 2.4 ANGSTROM RESOLUTION OF E. COLI SERINE HYDROXYMETHYLTRANSFERASE IN COMPLEX WITH GLYCINE AND 5-FORMYL TETRAHYDROFOLATE | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Scarsdale, J.N, Radaev, S, Kazanina, G, Schirch, V, Wright, H.T. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure at 2.4 A resolution of E. coli serine hydroxymethyltransferase in complex with glycine substrate and 5-formyl tetrahydrofolate.
J.Mol.Biol., 296, 2000
|
|
1I30
 
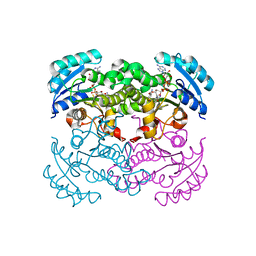 | | E. Coli Enoyl Reductase +NAD+SB385826 | | Descriptor: | 1,3,4,9-TETRAHYDRO-2-(HYDROXYBENZOYL)-9-[(4-HYDROXYPHENYL)METHYL]-6-METHOXY-2H-PYRIDO[3,4-B]INDOLE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Seefeld, M.A, Miller, W.H, Payne, D.J, Janson, C.A, Qiu, X. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitors of bacterial enoyl acyl carrier protein reductase (FabI): 2,9-disubstituted 1,2,3,4-tetrahydropyrido[3,4-b]indoles as potential antibacterial agents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
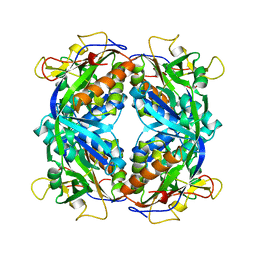
1M1P
 
 | | P21 crystal structure of the tetraheme cytochrome c3 from Shewanella oneidensis MR1 | | Descriptor: | HEME C, SULFATE ION, Small tetraheme cytochrome c | | Authors: | Leys, D, Meyer, T.E, Tsapin, A.I, Nealson, K.H, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 2002-06-20 | | Release date: | 2002-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures at atomic resolution reveal the novel concept of 'electron-harvesting' as a role for the small tetraheme cytochrome c
J.Biol.Chem., 277, 2002
|
|
1FTR
 
 | |
8FI4
 
 | |
8FI6
 
 | |
8FI5
 
 | |
5J63
 
 | | Crystal Structure of the N-terminal N-formyltransferase Domain (residues 1-306) of Escherichia coli Arna in Complex with UDP-Ara4N and Folinic Acid | | Descriptor: | (2R,3R,4S,5S)-5-amino-3,4-dihydroxytetrahydro-2H-pyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, (4aS,6S)-2-amino-6-{(E)-[(4-methylphenyl)imino]methyl}-4-oxo-4,6,7,8-tetrahydropteridine-5(4aH)-carbaldehyde, Bifunctional polymyxin resistance protein ArnA | | Authors: | Thoden, J.B, Genthe, N.A, Holden, H.M. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Escherichia coli ArnA N-formyltransferase domain in complex with N(5) -formyltetrahydrofolate and UDP-Ara4N.
Protein Sci., 25, 2016
|
|
6PVW
 
 | | RNase A in complex with cp4pA | | Descriptor: | 2,4,6,8-tetrahydroxy-1,3,5,7,2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~-tetroxatetraphosphocane-2,4,6,8-tetrone, 5'-O-[(R)-hydroxy{[(4R,8S)-4,6,8-trihydroxy-2,4,6,8-tetraoxo-1,3,5,7,2lambda~5~,4lambda~5~,6lambda~5~,8lambda~5~-tetroxatetraphosphocan-2-yl]oxy}phosphoryl]adenosine, Ribonuclease pancreatic | | Authors: | Windsor, I.W, Sheppard, S.M, Cummins, C.C, Raines, R.T. | | Deposit date: | 2019-07-21 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nucleoside Tetra- and Pentaphosphates Prepared Using a Tetraphosphorylation Reagent Are Potent Inhibitors of Ribonuclease A.
J.Am.Chem.Soc., 141, 2019
|
|
8CLR
 
 | | Integrated NMR/MD structure determination of a dynamic and thermodynamically stable CUUG RNA tetraloop | | Descriptor: | RNA hairpin with CUUG tetraloop | | Authors: | Oxenfarth, A, Kuemmerer, F, Bottaro, S, Schnieders, R, Pinter, G, Jonker, H.R.A, Fuertig, B, Richter, C, Blackledge, M, Lindorff-Larsen, K, Schwalbe, H. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Integrated NMR/Molecular Dynamics Determination of the Ensemble Conformation of a Thermodynamically Stable CUUG RNA Tetraloop.
J.Am.Chem.Soc., 145, 2023
|
|
4WTY
 
 | | Structure of the PTP-like myo-inositol phosphatase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ... | | Authors: | Bruder, L.M, Mosimann, S.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate
To Be Published
|
|
4WU3
 
 | | Structure of the PTP-like myo-inositol phosphatase from Mitsuokella multacida in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate | | Descriptor: | GLYCEROL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, MYO-INOSITOL PHOSPHOHYDROLASE, ... | | Authors: | Bruder, L.M, Mosimann, S.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate
To Be Published
|
|
4X6N
 
 | |
6RXB
 
 | | Crystal structure of TetR-Q116A from Acinetobacter baumannii AYE in complex with minocycline | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tam, H.K, Himpich, S, Pos, K.M. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding of Tetracyclines to Acinetobacter baumannii TetR Involves Two Arginines as Specificity Determinants
Front Microbiol, 2021
|
|
6OMK
 
 | |
4X6O
 
 | |
4X48
 
 | | Crystal structure of GluR2 ligand-binding core | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, N-{(3S,4S)-4-[4-(5-cyanothiophen-2-yl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide, ... | | Authors: | Pandit, J. | | Deposit date: | 2014-12-02 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Discovery and Characterization of the alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptor Potentiator N-{(3S,4S)-4-[4-(5-Cyano-2-thienyl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide (PF-04958242).
J.Med.Chem., 58, 2015
|
|
4X6P
 
 | |
8GZ2
 
 | | Cryo-EM structure of human NaV1.6/beta1/beta2-4,9-anhydro-tetrodotoxin | | Descriptor: | (1R,2S,3S,4R,5R,9S,11S,12S,14R)-7-amino-2,4,12-trihydroxy-2-(hydroxymethyl)-10,13,15-trioxa-6,8-diazapentacyclo[7.4.1.1~3,12~.0~5,11~.0~5,14~]pentadec-7-en-8-ium (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 8 subunit alpha, ... | | Authors: | Li, Y, Jiang, D. | | Deposit date: | 2022-09-24 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human Na V 1.6 channel reveals Na + selectivity and pore blockade by 4,9-anhydro-tetrodotoxin.
Nat Commun, 14, 2023
|
|
7SDD
 
 | | Structure of the PTP-like myo-inositol phosphatase from Legionella pneumophila str. Paris in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Myo-inositol phosphohydrolase | | Authors: | Cleland, C.P, Mosimann, S.C. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the PTP-like myo-inositol phosphatase from Legionella pneumophila str. Paris in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate
To Be Published
|
|
