1C2A
 
 | | CRYSTAL STRUCTURE OF BARLEY BBI | | Descriptor: | BOWMAN-BIRK TRYPSIN INHIBITOR | | Authors: | Song, H.K, Kim, Y.S, Yang, J.K, Moon, J, Lee, J.Y, Suh, S.W. | | Deposit date: | 1999-07-23 | | Release date: | 1999-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 A resolution.
J.Mol.Biol., 293, 1999
|
|
1MQ6
 
 | | Crystal Structure of 3-chloro-N-[4-chloro-2-[[(5-chloro-2-pyridinyl)amino]carbonyl]-6-methoxyphenyl]-4-[[(4,5-dihydro-2-oxazolyl)methylamino]methyl]-2-thiophenecarboxamide Complexed with Human Factor Xa | | Descriptor: | 3-CHLORO-N-[4-CHLORO-2-[[(5-CHLORO-2-PYRIDINYL)AMINO]CARBONYL]-6-METHOXYPHENYL]-4-[[(4,5-DIHYDRO-2-OXAZOLYL)METHYLAMINO]METHYL]-2-THIOPHENECARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X HEAVY CHAIN, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2002-09-13 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Two Potent Nonamidine Inhibitors Bound to Factor Xa
Biochemistry, 41, 2002
|
|
3ARZ
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with 2-(imidazolin-2-yl)-5-isothiocyanatobenzofuran | | Descriptor: | 2-(5-isothiocyanato-1-benzofuran-2-yl)-4,5-dihydro-1H-imidazole, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
8C3V
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-13 Fab and C1 nanobody | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, BA.2-13 heavy chain, BA.2-13 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-12-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
5QS5
 
 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury in complex with Z32400357 | | Descriptor: | 1-(3-methylbenzene-1-carbonyl)piperidine-4-carboxamide, CADMIUM ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Fernandez-Cid, A, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2C03
 
 | | GDP COMPLEX OF SRP GTPASE FFH NG DOMAIN | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ramirez, U.D, Preininger, A.M, Freymann, D.M. | | Deposit date: | 2005-08-25 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Nucleotide-Binding Flexibility in Ultrahigh-Resolution Structures of the Srp Gtpase Ffh
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5E9B
 
 | | Crystal structure of human heparanase in complex with HepMer M09S05a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, CHLORIDE ION, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2015-10-14 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural characterization of human heparanase reveals insights into substrate recognition.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5TDI
 
 | | Crystal structure of Cathepsin K with a covalently-linked inhibitor at 1.4 Angstrom resolution. | | Descriptor: | 4-fluoro-N-{1-[(Z)-iminomethyl]cyclopropyl}-N~2~-{(1S)-2,2,2-trifluoro-1-[4'-(methylsulfonyl)[1,1'-biphenyl]-4-yl]ethyl }-L-leucinamide, Cathepsin K | | Authors: | Law, S, Aguda, A, Nguyen, N, Brayer, G, Bromme, D. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of mouse cathepsin K structural elements that regulate the potency of odanacatib.
Biochem. J., 474, 2017
|
|
2VJ3
 
 | | Human Notch-1 EGFs 11-13 | | Descriptor: | CALCIUM ION, CHLORIDE ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1, ... | | Authors: | Johnson, S, Cordle, J, Tay, J.Z, Roversi, P, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Conserved Face of the Jagged/Serrate Dsl Domain is Involved in Notch Trans-Activation and Cis-Inhibition.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1ME4
 
 | |
4I87
 
 | | Crystal structure of TTR variant I84S in complex with CHF5074 at acidic pH | | Descriptor: | 1-(3',4'-dichloro-2-fluorobiphenyl-4-yl)cyclopropanecarboxylic acid, Transthyretin | | Authors: | Zanotti, G, Cendron, L, Folli, C, Florio, P, Imbimbo, B.P, Berni, R. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural evidence for native state stabilization of a conformationally labile amyloidogenic transthyretin variant by fibrillogenesis inhibitors.
Febs Lett., 587, 2013
|
|
1C1K
 
 | | BACTERIOPHAGE T4 GENE 59 HELICASE ASSEMBLY PROTEIN | | Descriptor: | BPT4 GENE 59 HELICASE ASSEMBLY PROTEIN, CHLORIDE ION, IRIDIUM ION | | Authors: | Mueser, T.C, Jones, C.E, Nossal, N.G, Hyde, C.C. | | Deposit date: | 1999-07-22 | | Release date: | 2000-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bacteriophage T4 gene 59 helicase assembly protein binds replication fork DNA. The 1.45 A resolution crystal structure reveals a novel alpha-helical two-domain fold.
J.Mol.Biol., 296, 2000
|
|
1WQ5
 
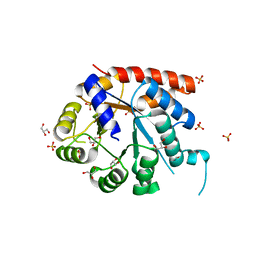 | | Crystal structure of tryptophan synthase alpha-subunit from Escherichia coli | | Descriptor: | GLYCEROL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Nishio, K, Morimoto, Y, Ishizuka, M, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-22 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Changes in the alpha-Subunit Coupled to Binding of the beta(2)-Subunit of Tryptophan Synthase from Escherichia coli: Crystal Structure of the Tryptophan Synthase alpha-Subunit Alon
Biochemistry, 44, 2005
|
|
3PJC
 
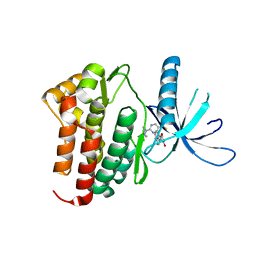 | | Crystal structure of JAK3 complexed with a potent ATP site inhibitor showing high selectivity within the Janus kinase family | | Descriptor: | 3-(1H-indol-3-yl)-4-[2-(4-oxopiperidin-1-yl)-5-(trifluoromethyl)pyrimidin-4-yl]-1H-pyrrole-2,5-dione, Tyrosine-protein kinase JAK3 | | Authors: | Tavares, G.A, Thoma, G, Zerwes, H.-G, Kroemer, M. | | Deposit date: | 2010-11-09 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Potent Janus Kinase 3 Inhibitor with High Selectivity within the Janus Kinase Family.
J.Med.Chem., 54, 2011
|
|
3PUA
 
 | | PHF2 Jumonji-NOG-Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-OXALYLGLYCINE, ... | | Authors: | Horton, J.R, Upadhyay, A.K, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2010-12-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for human PHF2 Jumonji domain interaction with metal ions.
J.Mol.Biol., 406, 2011
|
|
5U21
 
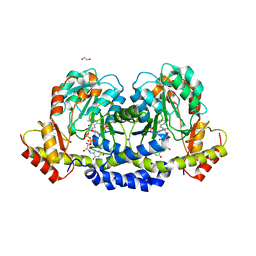 | | X-ray structure of the WlaRF aminotransferase from Campylobacter jejuni, K184A mutant in complex with TDP-Qui3N | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5QY0
 
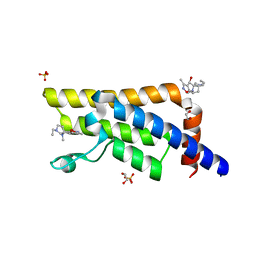 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with PC591 | | Descriptor: | (4R,4aS,7aS,9S)-6-ethyl-3,10-dimethyl-5,6,7,7a,8,9-hexahydro-4H-4a,9-epiminopyrrolo[3',4':5,6]cyclohepta[1,2-d][1,2]oxazol-4-ol, 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
5E98
 
 | |
5QY3
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment F2X-Entry B03a | | Descriptor: | 2-[(1~{S})-1-azanylpropyl]phenol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6J26
 
 | | Crystal structure of the branched-chain polyamine synthase from Thermococcus kodakarensis (Tk-BpsA) in complex with N4-bis(aminopropyl)spermidine and 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, FE (III) ION, N(4)-bis(aminopropyl)spermidine synthase, ... | | Authors: | Mizohata, E, Toyoda, M, Fujita, J, Inoue, T. | | Deposit date: | 2018-12-31 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The C-terminal flexible region of branched-chain polyamine synthase facilitates substrate specificity and catalysis.
Febs J., 286, 2019
|
|
4HXM
 
 | | Brd4 Bromodomain 1 complex with N-{3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)-5-[(THIOPHEN-2-YLSULFONYL)AMINO]PHENYL}BUTANAMIDE inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-{3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)-5-[(thiophen-2-ylsulfonyl)amino]phenyl}butanamide | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
7EW9
 
 | | GDP-bound KRAS G12D in complex with TH-Z816 | | Descriptor: | 7-(8-methylnaphthalen-1-yl)-4-[(2~{R})-2-methylpiperazin-1-yl]-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
1Q03
 
 | | Crystal structure of FGF-1, S50G/V51G mutant | | Descriptor: | Heparin-binding growth factor 1 | | Authors: | Kim, J, Blaber, M. | | Deposit date: | 2003-07-15 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Sequence swapping does not result in conformation swapping for the beta4/beta5 and beta8/beta9 beta-hairpin turns in human acidic fibroblast growth factor
Protein Sci., 14, 2005
|
|
1BMB
 
 | | GRB2-SH2 DOMAIN IN COMPLEX WITH KPFY*VNVEF (PKF270-974) | | Descriptor: | PROTEIN (GROWTH FACTOR RECEPTOR BOUND PROTEIN 2), PROTEIN (PKF270-974) | | Authors: | Rondeau, J.M, Zurini, M. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and conformational requirements for high-affinity binding to the SH2 domain of Grb2(1).
J.Med.Chem., 42, 1999
|
|
