1BAP
 
 | |
1APB
 
 | |
4DZL
 
 | | A de novo designed Coiled Coil CC-Tri | | Descriptor: | COILED-COIL PEPTIDE CC-TRI | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
4DZK
 
 | | A de novo designed Coiled Coil CC-Tri-N13 | | Descriptor: | CHLORIDE ION, COILED-COIL PEPTIDE CC-TRI-N13 | | Authors: | Bruning, M, Thomson, A.R, Zaccai, N.R, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2012-03-01 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A basis set of de novo coiled-coil Peptide oligomers for rational protein design and synthetic biology.
ACS Synth Biol, 1, 2012
|
|
4PRF
 
 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
6E7G
 
 | |
6ES4
 
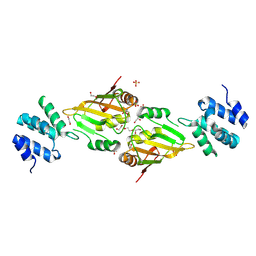 | | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Syncrip, ... | | Authors: | Hobor, F, Dallmann, A, Ball, N.J, Cicchini, C, Battistelli, C, Ogrodowicz, R.W, Christodoulou, E, Martin, S.R, Castello, A, Tripodi, M, Taylor, I.A, Ramos, A. | | Deposit date: | 2017-10-19 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets.
Nat Commun, 9, 2018
|
|
5H2L
 
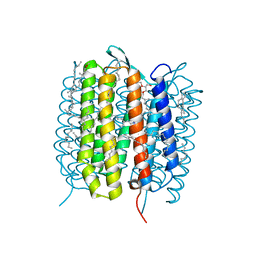 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 5.25 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2N
 
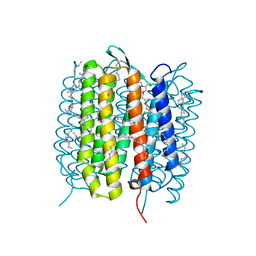 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 95.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2O
 
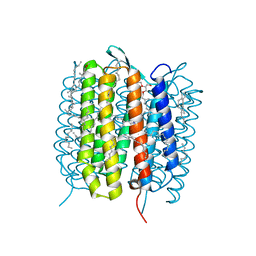 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 250 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
3N5N
 
 | |
3J4U
 
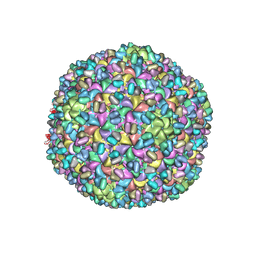 | | A new topology of the HK97-like fold revealed in Bordetella bacteriophage: non-covalent chainmail secured by jellyrolls | | Descriptor: | cementing protein, major capsid protein | | Authors: | Zhang, X, Guo, H, Jin, L, Czornyj, E, Hodes, A, Hui, W.H, Nieh, A.W, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2013-10-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution.
Elife, 2, 2013
|
|
2KJI
 
 | | A divergent ins protein in c. elegans structurally resemble insulin and activates the human insulin receptor | | Descriptor: | Probable insulin-like peptide beta-type 5 | | Authors: | Hua, Q.X, Nakarawa, S.H, Wilken, R, Ramos, R.R, Jia, W.H, Bass, J, Weiss, M.A. | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | A divergent INS protein in Caenorhabditis elegans structurally resembles human insulin and activates the human insulin receptor.
Genes Dev., 17, 2003
|
|
3RTR
 
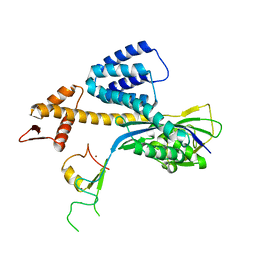 | | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, ZINC ION | | Authors: | Calabrese, M.F, Scott, D.C, Duda, D.M, Grace, C.R, Kurinov, I, Kriwacki, R.W, Schulman, B.A. | | Deposit date: | 2011-05-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases.
Nat.Struct.Mol.Biol., 18, 2011
|
|
4ZQX
 
 | | A revised partiality model and post-refinement algorithm for X-ray free-electron laser data | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Polyhedrin | | Authors: | Ginn, H.M, Brewster, A.S, Hattne, J, Evans, G, Wagner, A, Grimes, J, Sauter, N.K, Sutton, G, Stuart, D.I. | | Deposit date: | 2015-05-11 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A revised partiality model and post-refinement algorithm for X-ray free-electron laser data.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6CHA
 
 | |
3L56
 
 | | Crystal structure of the large c-terminal domain of polymerase basic protein 2 from influenza virus a/viet nam/1203/2004 (h5n1) | | Descriptor: | Polymerase PB2 | | Authors: | Staker, B.L, Edwards, T, Eric, S, Raymond, A, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-12-21 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biological and structural characterization of a host-adapting amino acid in influenza virus.
Plos Pathog., 6, 2010
|
|
4QNP
 
 | | Crystal structure of the 2009 pandemic H1N1 influenza virus neuraminidase with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wan, H.Q, Yang, H, Shore, D.A, Garten, R.J, Couzens, L, Gao, J, Jiang, L.L, Carney, P.J, Villanueva, J, Stevens, J, Eichelberger, M.C. | | Deposit date: | 2014-06-18 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers.
Nat Commun, 6, 2015
|
|
4R0Z
 
 | | A conserved phosphorylation switch controls the interaction between cadherin and beta-catenin in vitro and in vivo | | Descriptor: | FORMIC ACID, Protein humpback-2 | | Authors: | Choi, H.-J, Loveless, T, Lynch, A, Bang, I, Hardin, J, Weis, W.I. | | Deposit date: | 2014-08-03 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | A Conserved Phosphorylation Switch Controls the Interaction between Cadherin and beta-Catenin In Vitro and In Vivo
Dev.Cell, 33, 2015
|
|
4F04
 
 | | A Second Allosteric site in E. coli Aspartate Transcarbamoylase: R-state ATCase with UTP bound | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Peterson, A.W, Cockrell, G.M, Kantrowitz, E.R. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A second allosteric site in Escherichia coli aspartate transcarbamoylase.
Biochemistry, 51, 2012
|
|
11BG
 
 | | A POTENTIAL ALLOSTERIC SUBSITE GENERATED BY DOMAIN SWAPPING IN BOVINE SEMINAL RIBONUCLEASE | | Descriptor: | PROTEIN (BOVINE SEMINAL RIBONUCLEASE), SULFATE ION, URIDYLYL-2'-5'-PHOSPHO-GUANOSINE | | Authors: | Vitagliano, L, Adinolfi, S, Sica, F, Merlino, A, Zagari, A, Mazzarella, L. | | Deposit date: | 1999-03-11 | | Release date: | 1999-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A potential allosteric subsite generated by domain swapping in bovine seminal ribonuclease.
J.Mol.Biol., 293, 1999
|
|
4PAQ
 
 | | A conserved phenylalanine as relay between the 5 helix and the GDP binding region of heterotrimeric G protein | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, MAGNESIUM ION | | Authors: | Kaya, A.I, Lokits, A.D, Gilbert, J, Iverson, T.M, Meiler, J, Hamm, H.E. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Conserved Phenylalanine as a Relay between the alpha 5 Helix and the GDP Binding Region of Heterotrimeric Gi Protein alpha Subunit.
J.Biol.Chem., 289, 2014
|
|
3Q5U
 
 | |
3LT6
 
 | |
4JMR
 
 | | A unique spumavirus gag N-terminal domain with functional properties of orthoretroviral Matrix and Capsid | | Descriptor: | Env protein, Gag protein | | Authors: | Taylor, I.A, Goldstone, D.C, Flower, T.G, Ball, N.J. | | Deposit date: | 2013-03-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Unique Spumavirus Gag N-terminal Domain with Functional Properties of Orthoretroviral Matrix and Capsid.
Plos Pathog., 9, 2013
|
|
