7BDT
 
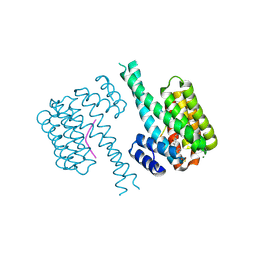 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1009 | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-(2-phenylimidazol-1-yl)benzaldehyde, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BGV
 
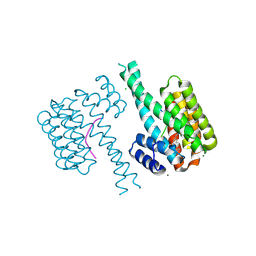 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1012 | | Descriptor: | 14-3-3 protein sigma, 3-methoxy-4-(2-phenylimidazol-1-yl)benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BDY
 
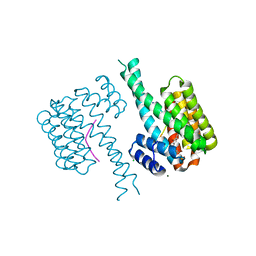 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2068B | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-[2-(5-bromanyl-2-fluoranyl-phenyl)imidazol-1-yl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2020-12-22 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BFW
 
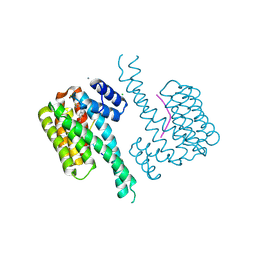 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2068A | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BG3
 
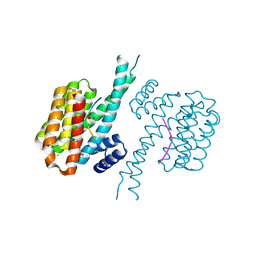 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound PC2046 | | Descriptor: | 1-(3-bromanyl-4-methyl-phenyl)-2-(2-bromophenyl)imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7BGW
 
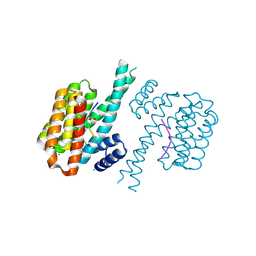 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1011 | | Descriptor: | 14-3-3 protein sigma, 4-(2-phenylimidazol-1-yl)naphthalene-1-carbaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-01-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
4GTF
 
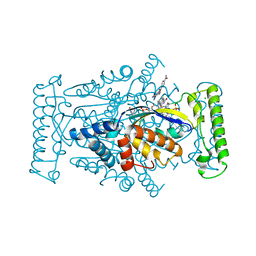 | | T. Maritima FDTS (H53A mutant) with FAD, dUMP and Folate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7SNG
 
 | | structure of G6PD-WT tetramer | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Allosteric role of a structural NADP + molecule in glucose-6-phosphate dehydrogenase activity.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6C14
 
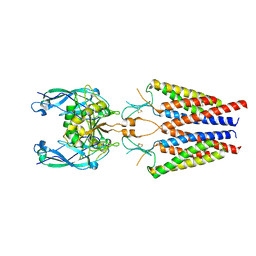 | |
4GT9
 
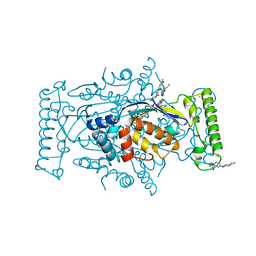 | | T. Maritima FDTS with FAD, dUMP and Folate. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mathews, I.I, Lesley, S.A, Kohen, A. | | Deposit date: | 2012-08-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7BO6
 
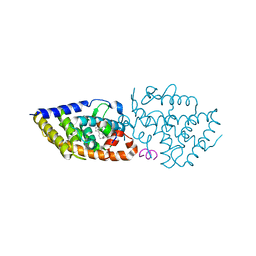 | | VDR complex with LCA derivative | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-01-24 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lithocholic acid-based design of noncalcemic vitamin D receptor agonists.
Bioorg.Chem., 111, 2021
|
|
7BK4
 
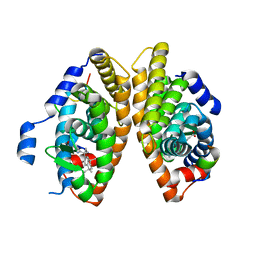 | | Crystal structure of RXRalpha ligand binding domain in complex with a fragment of the TIF2 coactivator | | Descriptor: | 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | le Maire, A, Bourguet, W. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J.Mol.Biol., 433, 2021
|
|
7BBT
 
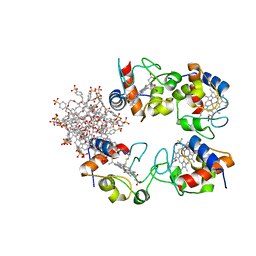 | | Structure of cytochrome c in complex with a p-benzyl-sulfonato-calix[8]arene-PEG pseudorotaxane | | Descriptor: | 4-[[49,50,51,52,53,54,55,56-octahydroxy-11,17,23,29,35,41,47-heptakis[(4-sulfonatophenyl)methyl]-5-nonacyclo[43.3.1.13,7.19,13.115,19.121,25.127,31.133,37.139,43]hexapentaconta-1(49),3,5,7(56),9,11,13(55),15,17,19(54),21,23,25(53),27,29,31(52),33,35,37(51),39,41,43(50),45,47-tetracosaenyl]methyl]benzenesulfonate, Cytochrome c iso-1, HEME C, ... | | Authors: | Mockler, N.M, Ramberg, K, Guagnini, F, Raston, C.L, Crowley, P.B. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.023 Å) | | Cite: | Noncovalent Protein-Pseudorotaxane Assembly Incorporating an Extended Arm Calix[8]arene with alpha-Helical Recognition Properties.
Cryst.Growth Des., 21, 2021
|
|
8XUQ
 
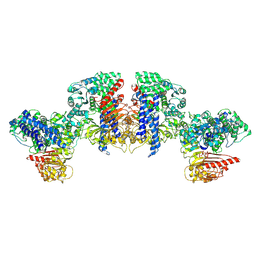 | | Cryo-EM structure of tomato NRC2 tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, NRC2 | | Authors: | Sun, Y, Ma, S.C, Chai, J.J. | | Deposit date: | 2024-01-14 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Oligomerization-mediated autoinhibition and cofactor binding of a plant NLR.
Nature, 2024
|
|
8E7O
 
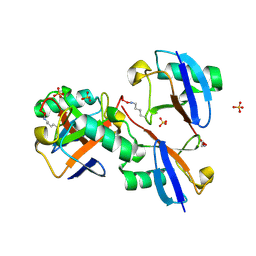 | | CRYSTAL STRUCTURE OF LYS48-LINKED TETRAUBIQUITIN | | Descriptor: | SULFATE ION, Ubiquitin | | Authors: | Lemma, B.E, Fushman, D. | | Deposit date: | 2022-08-24 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of selective recognition of Lys48-linked polyubiquitin by macrocyclic peptide inhibitors of proteasomal degradation.
Nat Commun, 14, 2023
|
|
7CTA
 
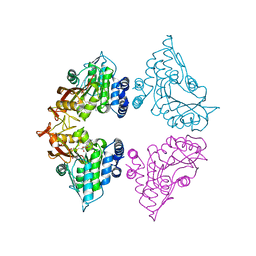 | | Crystal structure of Cx-SAM bound CmoB from Vibrio vulnificus | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, SULFATE ION, tRNA U34 carboxymethyltransferase | | Authors: | Kim, J, Jeong, S. | | Deposit date: | 2020-08-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural snapshots of CmoB in various states during wobble uridine modification of tRNA.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7UAL
 
 | |
6CO9
 
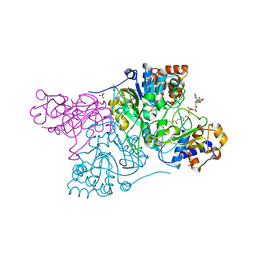 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB COCHEA-COA complex | | Descriptor: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W26
 
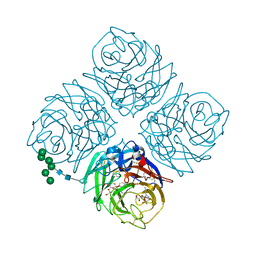 | | INFLUENZA VIRUS NEURAMINIDASE N9 IN COMPLEX WITH 4-DEOXYGENATED 2,3-DIFLUORO-N-ACETYLNEURAMINIC ACID | | Descriptor: | (2R,3R,5R,6R)-5-acetamido-2,3-bis(fluoranyl)-6-[(1R,2R)-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, (2~{R},3~{R},5~{R})-3-acetamido-5-fluoranyl-2-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]-2,3,4,5-tetrahydropyran-1-ium-6-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Mckimm-Breschkin, J, Barrett, S, Pilling, P, Hader, S, Watt, A.G. | | Deposit date: | 2017-06-05 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of Anti-Influenza Activity of 4-, 7-, 8- and 9-Deoxygenated 2,3-Difluoro- N-acetylneuraminic Acid Derivatives.
J. Med. Chem., 61, 2018
|
|
6COJ
 
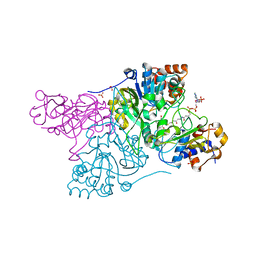 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB E105A COCHEA-COA complex | | Descriptor: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W2U
 
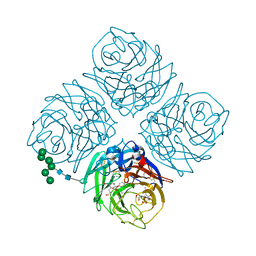 | | INFLUENZA VIRUS NEURAMINIDASE N9 IN COMPLEX WITH 7-DEOXYGENATED 2,3-DIFLUORO-N-ACETYLNEURAMINIC ACID | | Descriptor: | (2R,3R,4R,5R,6S)-5-acetamido-6-[(2S)-2,3-bis(oxidanyl)propyl]-2,3-bis(fluoranyl)-4-oxidanyl-oxane-2-carboxylic acid, (2~{S},3~{R},4~{R},5~{R})-3-acetamido-2-[(2~{S})-2,3-bis(oxidanyl)propyl]-5-fluoranyl-4-oxidanyl-2,3,4,5-tetrahydropyran-1-ium-6-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Streltsov, V.A, Mckimm-Breschkin, J, Barrett, S, Pilling, P, Hader, S, Watt, A.G. | | Deposit date: | 2017-06-06 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Anti-Influenza Activity of 4-, 7-, 8- and 9-Deoxygenated 2,3-Difluoro- N-acetylneuraminic Acid Derivatives.
J. Med. Chem., 61, 2018
|
|
5WDG
 
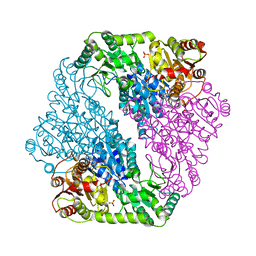 | | Acetolactate Synthase from Klebsiella pneumoniae in Complex with a Reaction Intermediate | | Descriptor: | (2S,3S)-2,3-dihydroxy-3-[(7S,8R,9aS)-8-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-2,7-dimethyl-5,7,8,10-tetrahydro-9aH-pyrimido[4,5-d][1,3]thiazolo[3,2-a]pyrimidin-9a-yl]-2-methylbutanoic acid, Acetolactate synthase, catabolic, ... | | Authors: | Latta, A.J, Andrews, F.H, McLeish, M.J. | | Deposit date: | 2017-07-05 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Acetolactate Synthase from Klebsiella pneumoniae in Complex with Mechanism-Based Inhibitor
To Be Published
|
|
5D7J
 
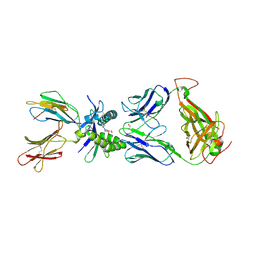 | | Structure of human MR1-5-OP-RU in complex with human MAIT M33.64(Y95alphaF) TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Keller, A.N, Woolley, R.E, Rossjohn, J. | | Deposit date: | 2015-08-14 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Diversity of T Cells Restricted by the MHC Class I-Related Molecule MR1 Facilitates Differential Antigen Recognition.
Immunity, 44, 2016
|
|
3SRK
 
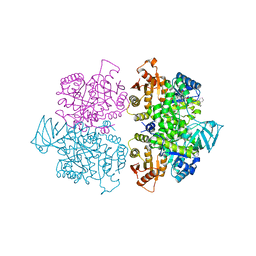 | | A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, POTASSIUM ION, Pyruvate kinase, ... | | Authors: | Morgan, H.P, Walsh, M, Blackburn, E.A, Wear, M.A, Boxer, M, Shen, M, McNae, I.W, Michels, P.A.M, Auld, D.S, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A new class of suicide inhibitor blocks nucleotide binding to pyruvate kinase
To be Published
|
|
4CSA
 
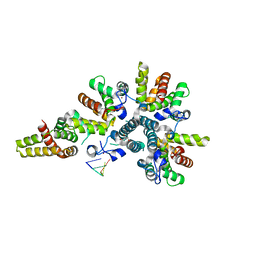 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer bound to a DNA 4-mer | | Descriptor: | 5'-D(*AP*GP*TP*TP*AP)-3', GLYCEROL, M2-1, ... | | Authors: | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | Deposit date: | 2014-03-05 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
