3OGP
 
 | | Crystal Structure of 6s-98S FIV Protease with Darunavir bound | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, DIMETHYL SULFOXIDE, FIV Protease | | Authors: | Lin, Y.-C, Perryman, A.L, Elder, J.H, Stout, C.D. | | Deposit date: | 2010-08-17 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for drug and substrate specificity exhibited by FIV encoding a chimeric FIV/HIV protease.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4QEL
 
 | | Crystal Structure of Benzoylformate Decarboxylase Mutant H70A | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Andrews, F.H, Rogers, M.P, Brodkin, H.R, McLeish, M.J. | | Deposit date: | 2014-05-16 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Structural investigation of benzoylformate decarboxylase active site variants
To be Published
|
|
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | Authors: | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2014-05-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
3BRG
 
 | | CSL (RBP-Jk) bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*DAP*DAP*DTP*DCP*DTP*DTP*DTP*DCP*DCP*DCP*DAP*DCP*DAP*DGP*DT)-3'), DNA (5'-D(*DTP*DTP*DAP*DCP*DTP*DGP*DTP*DGP*DGP*DGP*DAP*DAP*DAP*DGP*DA)-3'), ... | | Authors: | Friedmann, D.R, Kovall, R.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RAM-induced Allostery Facilitates Assembly of a Notch Pathway Active Transcription Complex.
J.Biol.Chem., 283, 2008
|
|
4LXX
 
 | | Crystal structure WlaRD, a sugar 3N-formyl transferase in the presence of dTDP-Fuc3NFo and 5-N-Formyl-THF | | Descriptor: | (2R,3R,4S,5R,6R)-4-(formylamino)-3,5-dihydroxy-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | thoden, J.B, goneau, M.-F, gilbert, M, holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4QG8
 
 | | crystal structure of PKM2-K305Q mutant | | Descriptor: | GLYCEROL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
4LVB
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-[4-(acetylamino)phenyl]cyclopropanecarboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.836 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1WGN
 
 | | Solution Structure of UBA domain of Human Ubiquitin Associated Protein 1 (UBAP1) | | Descriptor: | ubiquitin associated protein | | Authors: | Zhao, C, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of UBA domain of Human Ubiquitin Associated Protein 1 (UBAP1)
To be Published
|
|
3BYU
 
 | | co-crystal structure of Lck and aminopyrimidine reverse amide 23 | | Descriptor: | 2-methyl-N-{4-methyl-3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-5-yl)carbamoyl]phenyl}-3-(trifluoromethyl)benzamide, Proto-oncogene tyrosine-protein kinase LCK | | Authors: | Huang, X. | | Deposit date: | 2008-01-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of aminopyrimidine amides as potent, selective inhibitors of lymphocyte specific kinase: synthesis, structure-activity relationships, and inhibition of in vivo T cell activation.
J.Med.Chem., 51, 2008
|
|
4HM9
 
 | | Crystal structure of full-length human catenin-beta-like 1 | | Descriptor: | Beta-catenin-like protein 1 | | Authors: | Du, Z, Huang, X, Wang, G, Wu, Y. | | Deposit date: | 2012-10-18 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | The structure of full-length human CTNNBL1 reveals a distinct member of the armadillo-repeat protein family.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZKA
 
 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA D280N IN THE METAL-BOUND, OPEN STATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | Deposit date: | 2013-01-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
3ZMM
 
 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | 5-FLUORO-4-[(1S)-1-(5-FLUOROPYRIMIDIN-2-YL)ETHOXY]-N-(5-METHYL-1H-PYRAZOL-3-YL)-6-MORPHOLINO-PYRIMIDIN-2-AMINE, ACETYL GROUP, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J, Green, I, Pollard, H, Howard, T, Mott, R. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Novel Jak2-Stat Pathway Inhibitors with Extended Residence Time on Target.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HSP
 
 | |
2GAN
 
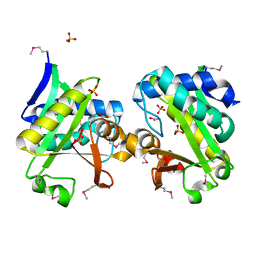 | | Crystal Structure of a Putative Acetyltransferase from Pyrococcus horikoshii, Northeast Structural Genomics Target JR32. | | Descriptor: | 1,2-ETHANEDIOL, 182aa long hypothetical protein, SULFATE ION | | Authors: | Forouhar, F, Abashidze, M, Jayaraman, S, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-09 | | Release date: | 2006-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of a Putative Acetyltransferase from Pyrococcus horikoshii, Northeast Structural Genomics Target JR32.
To be Published
|
|
1BUE
 
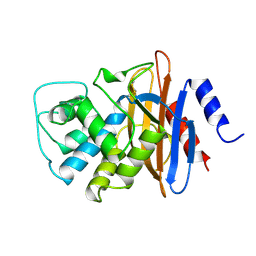 | | NMC-A CARBAPENEMASE FROM ENTEROBACTER CLOACAE | | Descriptor: | PROTEIN (IMIPENEM-HYDROLYSING BETA-LACTAMASE) | | Authors: | Swaren, P, Maveyraud, L, Cabantous, S, Pedelacq, J.D, Mourey, L, Frere, J.M, Samama, J.P. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | X-ray analysis of the NMC-A beta-lactamase at 1.64-A resolution, a class A carbapenemase with broad substrate specificity.
J.Biol.Chem., 273, 1998
|
|
4I9J
 
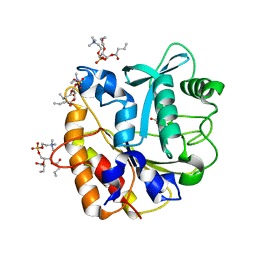 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to diC4PC | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | Deposit date: | 2012-12-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
5SAZ
 
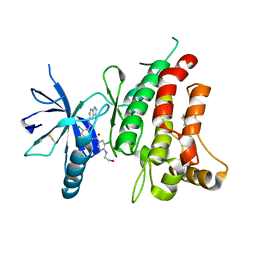 | | DDR1, 3-chloro-N-[4-chloro-3-(1H-pyrrolo[2,3-b]pyridin-5-ylcarbamoyl)phenyl]-4-(2-hydroxyethylamino)benzamide, 1.802A, P212121, Rfree=22.2% | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-5-{3-chloro-4-[(2-hydroxyethyl)amino]benzamido}-N-(1H-pyrrolo[2,3-b]pyridin-5-yl)benzamide, CHLORIDE ION, ... | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SAV
 
 | | DDR1, N-[2-[3-(2-aminopyrimidin-5-yl)oxyphenyl]ethyl]-3-(trifluoromethoxy)benzamide, 1.760A, P212121, Rfree=23.5% | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(2-{3-[(2-aminopyrimidin-5-yl)oxy]phenyl}ethyl)-3-(trifluoromethoxy)benzamide | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
4IAA
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with thioridazine | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Li, W, Xie, Y, Huang, N. | | Deposit date: | 2012-12-06 | | Release date: | 2013-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of Ser/Thr kinase Pim1 in complex with thioridazine
To be Published
|
|
5SAX
 
 | | DDR1, 2-[3-(2-pyridin-3-ylethynyl)phenyl]-N-[3-(trifluoromethyl)phenyl]acetamide, 1.902A, second P212121 form, Rfree=25.4%, second form | | Descriptor: | 2-{3-[(pyridin-3-yl)ethynyl]phenyl}-N-[3-(trifluoromethyl)phenyl]acetamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
5SAW
 
 | | DDR1, 2-[3-(2-pyridin-3-ylethynyl)phenyl]-N-[3-(trifluoromethyl)phenyl]acetamide, 1.601A, P212121, Rfree=22.6% | | Descriptor: | 2-{3-[(pyridin-3-yl)ethynyl]phenyl}-N-[3-(trifluoromethyl)phenyl]acetamide, Epithelial discoidin domain-containing receptor 1, IODIDE ION | | Authors: | Stihle, M, Richter, H, Benz, J, Hochstrasser, R, Rudolph, M.G. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal Structure of a DDR1 complex
To be published
|
|
1P48
 
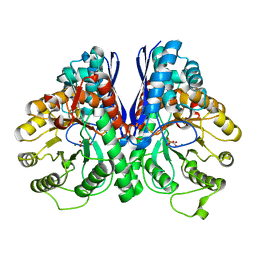 | | REVERSE PROTONATION IS THE KEY TO GENERAL ACID-BASE CATALYSIS IN ENOLASE | | Descriptor: | Enolase 1, MAGNESIUM ION, PHOSPHOENOLPYRUVATE | | Authors: | Sims, P.A, Larsen, T.M, Poyner, R.R, Cleland, W.W, Reed, G.H. | | Deposit date: | 2003-04-21 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reverse protonation is the key to general acid-base catalysis in enolase
Biochemistry, 42, 2003
|
|
1WS1
 
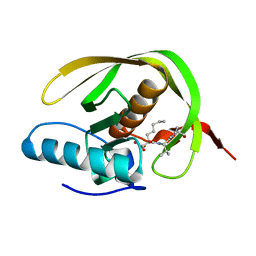 | |
1IU6
 
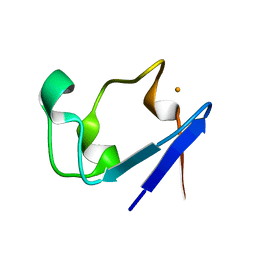 | | Neutron Crystal Structure of the rubredoxin mutant from Pyrococcus Furiosus | | Descriptor: | FE (III) ION, rubredoxin | | Authors: | Chatake, T, Kurihara, K, Tanaka, I, Tsyba, I, Bau, R, Jenney, F.E, Adams, M.W.W, Niimura, N. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (1.6 Å) | | Cite: | A neutron crystallographic analysis of a rubredoxin mutant at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3ZK7
 
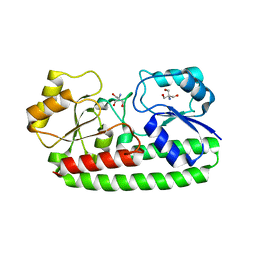 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA IN THE METAL-FREE, OPEN STATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | Deposit date: | 2013-01-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
