2HQ7
 
 | |
3GE5
 
 | |
3WRX
 
 | | Crystal structure of helicase complex 1 | | Descriptor: | CESIUM ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2C0Z
 
 | | The 1.6 A resolution crystal structure of NovW: a 4-keto-6-deoxy sugar epimerase from the novobiocin biosynthetic gene cluster of Streptomyces spheroides | | Descriptor: | 1,2-ETHANEDIOL, NOVW, SULFATE ION | | Authors: | Jakimowicz, P, Tello, M, Freel-Meyers, C.L, Walsh, C.T, Buttner, M.J, Field, R.A, Lawson, D.M. | | Deposit date: | 2005-09-09 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A Resolution Crystal Structure of Novw: A 4-Keto-6-Deoxy Sugar Epimerase from the Novobiocin Biosynthetic Gene Cluster of Streptomyces Spheroides
Proteins, 63, 2006
|
|
3G68
 
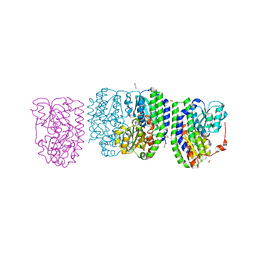 | |
4QJ8
 
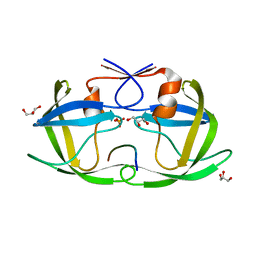 | |
3RPF
 
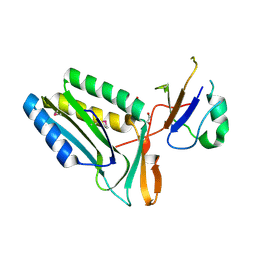 | | Protein-protein complex of subunit 1 and 2 of Molybdopterin-converting factor from Helicobacter pylori 26695 | | Descriptor: | 1,2-ETHANEDIOL, Molybdopterin converting factor, subunit 1 (MoaD), ... | | Authors: | Nocek, B, Stein, A, Marshall, N, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-26 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein-protein complex of subunit 1 and 2 of Molybdopterin-converting factor from Helicobacter pylori 26695
TO BE PUBLISHED
|
|
3ON6
 
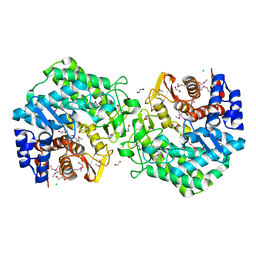 | |
3RU1
 
 | |
3I10
 
 | |
1BWO
 
 | |
3CWR
 
 | |
3CXW
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand I | | Descriptor: | (4R)-7,8-dichloro-1',9-dimethyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, CHLORIDE ION, Pimtide peptide, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Huber, K, Bracher, F, Pike, A.C.W, Roos, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 7,8-Dichloro-1-oxo-beta-carbolines as a Versatile Scaffold for the Development of Potent and Selective Kinase Inhibitors with Unusual Binding Modes
J.Med.Chem., 55, 2012
|
|
3ZSX
 
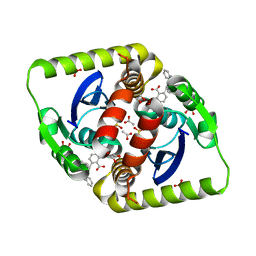 | | Small molecule inhibitors of the LEDGF site of HIV type 1 integrase identified by fragment screening and structure based drug design | | Descriptor: | 1,2-ETHANEDIOL, 5-({[2-(benzylcarbamoyl)benzyl](prop-2-en-1-yl)amino}methyl)-1,3-benzodioxole-4-carboxylate, ACETATE ION, ... | | Authors: | Peat, T.S, Newman, J, Rhodes, D.I, Vandergraaff, N, Le, G, Jones, E.D, Smith, J.A, Coates, J.A.V, Thienthong, N, Dolezal, O, Ryan, J.H, Savage, G.P, Francis, C.L, Deadman, J.J. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small Molecule Inhibitors of the Ledgf Site of Human Immunodeficiency Virus Integrase Identified by Fragment Screening and Structure Based Design.
Plos One, 7, 2012
|
|
1C3W
 
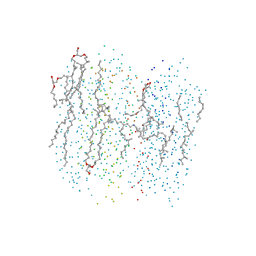 | | BACTERIORHODOPSIN/LIPID COMPLEX AT 1.55 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN (GROUND STATE WILD TYPE "BR"), ... | | Authors: | Luecke, H. | | Deposit date: | 1999-07-28 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of bacteriorhodopsin at 1.55 A resolution.
J.Mol.Biol., 291, 1999
|
|
4ZMJ
 
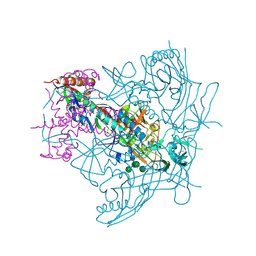 | | Crystal Structure of Ligand-Free BG505 SOSIP.664 HIV-1 Env Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2015-05-04 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3OS7
 
 | |
3D4E
 
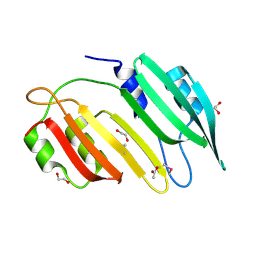 | |
3FSG
 
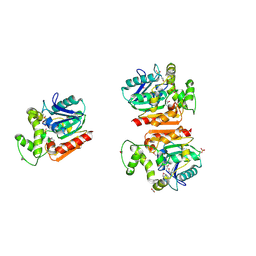 | | Crystal structure of alpha/beta superfamily hydrolase from Oenococcus oeni PSU-1 | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta superfamily hydrolase, CHLORIDE ION, ... | | Authors: | Nocek, B, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-09 | | Release date: | 2009-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alpha/beta superfamily hydrolase from Oenococcus oeni PSU-1
To be Published
|
|
1HEG
 
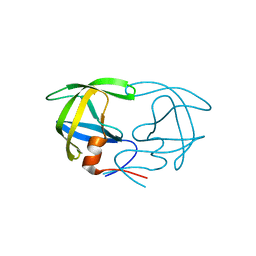 | | The crystal structures at 2.2 angstroms resolution of hydroxyethylene-based inhibitors bound to human immunodeficiency virus type 1 protease show that the inhibitors are present in two distinct orientations | | Descriptor: | HIV-1 PROTEASE, methyl N-{(4S,5S)-5-[(L-alanyl-L-alanyl)amino]-4-hydroxy-6-phenylhexanoyl}-L-valyl-L-valinate | | Authors: | Murthy, K, Winborne, E.L, Minnich, M.D, Culp, J.S, Debouck, C. | | Deposit date: | 1992-09-21 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures at 2.2-A resolution of hydroxyethylene-based inhibitors bound to human immunodeficiency virus type 1 protease show that the inhibitors are present in two distinct orientations.
J.Biol.Chem., 267, 1992
|
|
1TJP
 
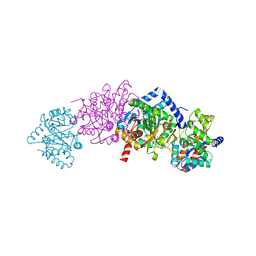 | | Crystal Structure Of Wild-Type Tryptophan Synthase Complexed With 1-[(2-hydroxylphenyl)amino]3-glycerolphosphate | | Descriptor: | 1-[(2-HYDROXYLPHENYL)AMINO]3-GLYCEROLPHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Kulik, V, Hartmann, E, Weyand, M, Frey, M, Gierl, A, Niks, D, Dunn, M.F, Schlichting, I. | | Deposit date: | 2004-06-07 | | Release date: | 2005-12-27 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | On the structural basis of the catalytic mechanism and the regulation of the alpha subunit of tryptophan synthase from Salmonella typhimurium and BX1 from maize, two evolutionarily related enzymes.
J.Mol.Biol., 352, 2005
|
|
3RVI
 
 | |
7DCZ
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S)-2-amino-4-methyl-4H-1,3-thiazin-4-yl]-4- fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Koriyama, Y, Hori, A, Ito, H, Yonezawa, S, Baba, Y, Tanimoto, N, Ueno, T, Yamamoto, S, Yamamoto, T, Asada, N, Morimoto, K, Einaru, S, Sakai, K, Kanazu, T, Matsuda, A, Yamaguchi, Y, Oguma, T, Timmers, M, Tritsmans, L, Kusakabe, K.I, Kato, A, Sakaguchi, G. | | Deposit date: | 2020-10-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Atabecestat (JNJ-54861911): A Thiazine-Based beta-Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor Advanced to the Phase 2b/3 EARLY Clinical Trial.
J.Med.Chem., 64, 2021
|
|
3RTM
 
 | |
1LRA
 
 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
