3NBP
 
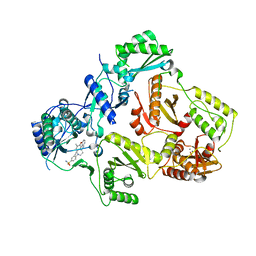 | | HIV-1 reverse transcriptase with aminopyrimidine inhibitor 2 | | Descriptor: | 4-(4-{[4-(4-cyano-2,6-dimethylphenoxy)pyrimidin-2-yl]amino}piperidin-1-yl)benzenesulfonamide, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Harris, S.F, Villasenor, A.G. | | Deposit date: | 2010-06-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of piperidin-4-yl-aminopyrimidines as HIV-1 reverse transcriptase inhibitors. N-benzyl derivatives with broad potency against resistant mutant viruses.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1FO2
 
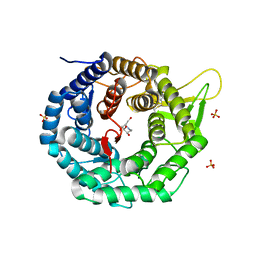 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE IN COMPLEX WITH 1-DEOXYMANNOJIRIMYCIN | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, ALPHA1,2-MANNOSIDASE, CALCIUM ION, ... | | Authors: | Vallee, F, Karaveg, K, Moremen, K.W, Herscovics, A, Howell, P.L. | | Deposit date: | 2000-08-24 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
5CPS
 
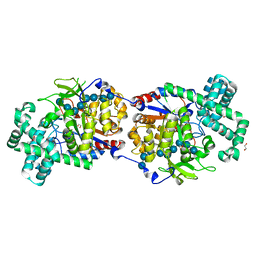 | | Disproportionating enzyme 1 from Arabidopsis - maltotriose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
3SE8
 
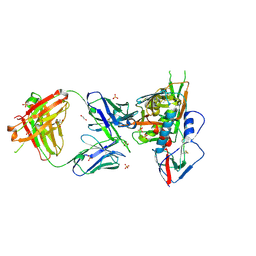 | | Crystal structure of broadly and potently neutralizing antibody VRC03 in complex with HIV-1 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwong, P.D, Zhou, T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
1KRE
 
 | | STRUCTURE OF P. CITRINUM ALPHA 1,2-MANNOSIDASE REVEALS THE BASIS FOR DIFFERENCES IN SPECIFICITY OF THE ER AND GOLGI CLASS I ENZYMES | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lobsanov, Y.D, Vallee, F, Imberty, A, Yoshida, T, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J.Biol.Chem., 277, 2002
|
|
4ICL
 
 | | HIV-1 reverse transcriptase with bound fragment at the incoming dNTP binding site | | Descriptor: | 4-(4-methylpiperazin-1-yl)benzoic acid, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-10 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4P7R
 
 | | Structure of Escherichia coli PgaB C-terminal domain in complex with a poly-beta-1,6-N-acetyl-D-glucosamine (PNAG) hexamer | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4OBD
 
 | |
1ZAI
 
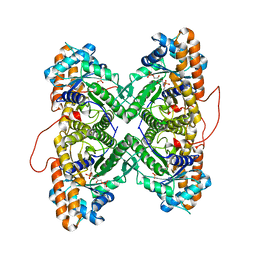 | | Fructose-1,6-bisphosphate Schiff base intermediate in FBP aldolase from rabbit muscle | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
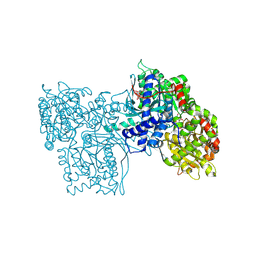
3GPB
 
 | |
3GUR
 
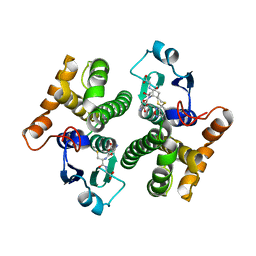 | | Crystal Structure of mu class glutathione S-transferase (GSTM2-2) in complex with glutathione and 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase Mu 2, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
1PMJ
 
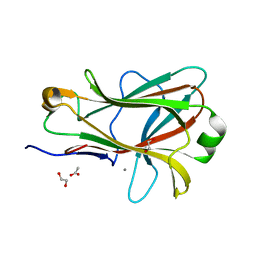 | | Crystal structure of Caldicellulosiruptor saccharolyticus CBM27-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Roske, Y, Sunna, A, Heinemann, U. | | Deposit date: | 2003-06-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution crystal structures of Caldicellulosiruptor strain Rt8B.4 carbohydrate-binding module CBM27-1 and its complex with mannohexaose.
J.Mol.Biol., 340, 2004
|
|
4EQ0
 
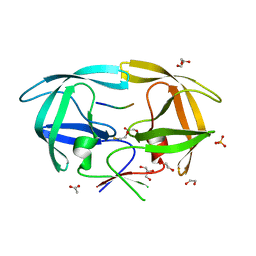 | | Crystal Structure of inactive single chain variant of HIV-1 Protease in Complex with the substrate p2-NC | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
4PUO
 
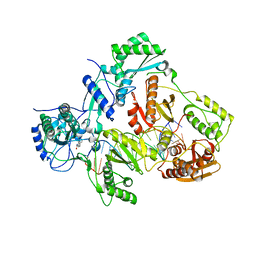 | | Crystal structure of HIV-1 reverse transcriptase in complex with RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*UP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-03-13 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
3D1R
 
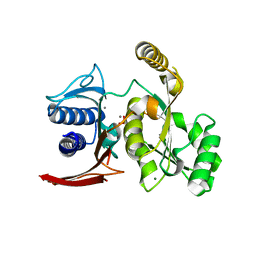 | | Structure of E. coli GlpX with its substrate fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A, Skarina, T, Dong, A, Brown, G, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3G2L
 
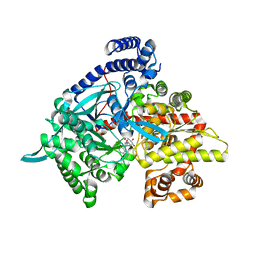 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazoles in complex with glycogen phosphorylase | | Descriptor: | 1-beta-D-glucopyranosyl-4-naphthalen-1-yl-1H-1,2,3-triazole, Glycogen phosphorylase, muscle form | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
4IFV
 
 | | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-Ray Crystallographic Fragment Screening | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, Exoribonuclease H, ... | | Authors: | Bauman, J.D, Patel, D, Fromer, M, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
3QD0
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with (2R,5S)-1-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-N-phenyl-3-piperidinecarboxamide | | Descriptor: | (3S,6R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-6-methyl-N-phenylpiperidine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3TOG
 
 | | HIV-1 Protease - Epoxydic Inhibitor Complex (pH 9 - Monoclinic Crystal form P21) | | Descriptor: | (S)-N-((2S,3S,4R,5R)-4-amino-3,5-dihydroxy-1,6-diphenylhexan-2-yl)-3-methyl-2-(2-phenoxyacetamido)butanamide, DIMETHYL SULFOXIDE, Gag-Pol polyprotein | | Authors: | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | Deposit date: | 2011-09-05 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Developing HIV-1 Protease Inhibitors through Stereospecific Reactions in Protein Crystals.
Molecules, 21, 2016
|
|
1J4E
 
 | | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE COVALENTLY BOUND TO THE SUBSTRATE DIHYDROXYACETONE PHOSPHATE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE A | | Authors: | Choi, K.H, Shi, J, Hopkins, C.E, Tolan, D.R, Allen, K.N. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Snapshots of catalysis: the structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate.
Biochemistry, 40, 2001
|
|
4GAE
 
 | | Crystal structure of plasmodium dxr in complex with a pyridine-containing inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, ... | | Authors: | Diao, J, Xue, J, Cai, G, Deng, L, Song, Y. | | Deposit date: | 2012-07-25 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antimalarial and Structural Studies of Pyridine-containing Inhibitors of 1-Deoxyxylulose-5-phosphate Reductoisomerase.
ACS Med Chem Lett, 4, 2013
|
|
4IG0
 
 | | HIV-1 reverse transcriptase with bound fragment at the 507 site | | Descriptor: | 2-({[2-(3,4-dihydroquinolin-1(2H)-yl)-2-oxoethyl](methyl)amino}methyl)quinazolin-4(1H)-one, DIMETHYL SULFOXIDE, P51 RT, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
1H5U
 
 | | THE 1.76 A RESOLUTION CRYSTAL STRUCTURE OF GLYCOGEN PHOSPHORYLASE B COMPLEXED WITH GLUCOSE AND CP320626, A POTENTIAL ANTIDIABETIC DRUG | | Descriptor: | 5-CHLORO-1H-INDOLE-2-CARBOXYLIC ACID [1-(4-FLUOROBENZYL)-2-(4-HYDROXYPIPERIDIN-1YL)-2-OXOETHYL]AMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Archontis, G. | | Deposit date: | 2001-05-25 | | Release date: | 2001-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The 1.76 A Resolution Crystal Structure of Glycogen Phosphorylase B Complexed with Glucose, and Cp320626, a Potential Antidiabetic Drug
Bioorg.Med.Chem., 10, 2002
|
|
1RTI
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 1-(2-HYDROXYETHYLOXYMETHYL)-6-PHENYL THIOTHYMINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
3G2H
 
 | | Crystal structure of 1-(beta-D-glucopyranosyl)-4-substituted-1,2,3-triazoles in complex with glycogen phosphorylase | | Descriptor: | 1-beta-D-glucopyranosyl-4-phenyl-1H-1,2,3-triazole, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Chrysina, E.D, Bokor, E, Alexacou, K.-M, Charavgi, M.-D, Oikonomakos, G.N, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2009-01-31 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Amide-1,2,3-triazole bioisosterism: the glycogen phosphorylase case
Tetrahedron: Asymmetry, 20, 2009
|
|
