5RF6
 
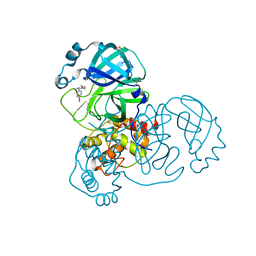 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1348371854 | | Descriptor: | 3C-like proteinase, 5-(1,4-oxazepan-4-yl)pyridine-2-carbonitrile, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3WD1
 
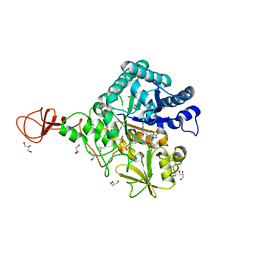 | | Serratia marcescens Chitinase B complexed with syn-triazole inhibitor | | Descriptor: | Chitinase B, GLYCEROL, N,N-dibenzyl-N~5~-[N-(methylcarbamoyl)carbamimidoyl]-N~2~-{[5-({[(E)-(quinolin-4-ylmethylidene)amino]oxy}methyl)-1H-1,2,3-triazol-1-yl]acetyl}-L-ornithinamide, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5RFK
 
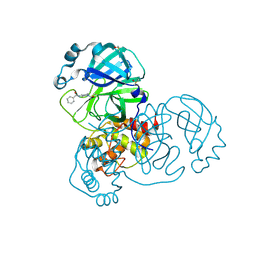 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102575 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(1-acetylpiperidin-4-yl)benzamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RG2
 
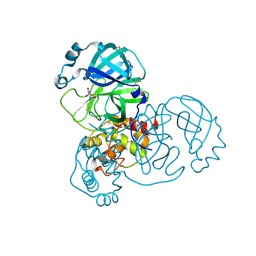 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with NCL-00025058 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N~2~-acetyl-N-prop-2-en-1-yl-D-allothreoninamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4O8N
 
 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, in the apoprotein form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, CALCIUM ION, ... | | Authors: | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | Deposit date: | 2013-12-28 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6476 Å) | | Cite: | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
5RGI
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z369936976 (Mpro-x0397) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N'-cyclopropyl-N-methyl-N-[(5-methyl-1,2-oxazol-3-yl)methyl]urea | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RGM
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102142 (Mpro-x0708) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N'-acetyl-4,5,6,7-tetrahydro-1-benzothiophene-2-carbohydrazide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-04-07 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
2R1S
 
 | |
2QJC
 
 | | Crystal structure of a putative diadenosine tetraphosphatase | | Descriptor: | Diadenosine tetraphosphatase, putative, MANGANESE (II) ION, ... | | Authors: | Sugadev, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-06 | | Release date: | 2007-07-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
5RFB
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1271660837 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1-methyl-1H-1,2,3-triazol-4-yl)methyl]ethanamine | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RHX
 
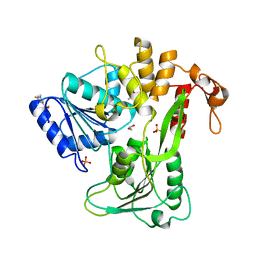 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z1324080698 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-fluoro-5-methylbenzene-1-sulfonamide, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GC6
 
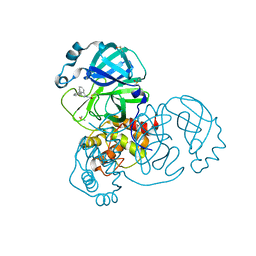 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with NIR-THE-c331be7a-6 (Mpro-x10513) | | Descriptor: | 1-[(4R)-4-(3-methylphenyl)-3,4-dihydroisoquinolin-2(1H)-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5RHM
 
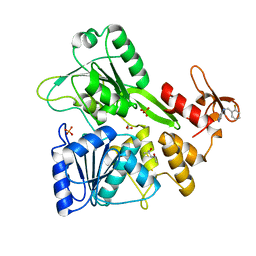 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z1454310449 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, N-[(2-fluorophenyl)methyl]-1H-pyrazol-4-amine, ... | | Authors: | Godoy, A.S, Mesquita, N.C.M.R, Oliva, G. | | Deposit date: | 2020-05-25 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7GBO
 
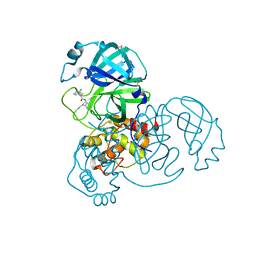 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAN-GHE-83b26c96-20 (Mpro-x10392) | | Descriptor: | 1-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)cyclopropane-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4OGP
 
 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2001-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
7KAN
 
 | | Cryo-EM structure of the Sec complex from T. lanuginosus, Sec62-lacking mutant (Delta Sec62) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Protein transport channel Sec61 complex, alpha subunit (Sec61), ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-02-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7GAZ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DUN-NEW-f8ce3686-14 (Mpro-x10049) | | Descriptor: | 1-{2-[(methanesulfonyl)amino]ethyl}-1,2,3,4-tetrahydroquinoline-7-sulfonamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4GVS
 
 | | X-ray structure of the Archaeoglobus fulgidus methenyl-tetrahydromethanopterin cyclohydrolase in complex with N5-formyl-tetrahydromethanopterin | | Descriptor: | 1-[4-({(1R)-1-[(6S,7R)-2-amino-5-formyl-7-methyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]ethyl}amino)phenyl]-1-deoxy-5 -O-{5-O-[(R)-{[(1R)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, Methenyltetrahydromethanopterin cyclohydrolase | | Authors: | Upadhyay, V, Demmer, U, Warkentin, E, Moll, J, Shima, S, Ermler, U. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and catalytic mechanism of N(5),N(10)-methenyl-tetrahydromethanopterin cyclohydrolase.
Biochemistry, 51, 2012
|
|
7GAW
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-e194df51-1 (SARS2_MproA-x0862) | | Descriptor: | (4S)-6-chloro-2-[(1-cyanocyclopropyl)methanesulfonyl]-N-(isoquinolin-4-yl)-1,2,3,4-tetrahydroisoquinoline-4-carboxamide, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4MN3
 
 | | Chromodomain antagonists that target the polycomb-group methyllysine reader protein Chromobox homolog 7 (CBX7) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Chromobox protein homolog 7, ... | | Authors: | Chakravarthi, S, Daze, K, Douglas, S, Quon, T, Dev, A, Peng, F, Heller, M, Boulanger, M.J, Wulff, J, Hof, F. | | Deposit date: | 2013-09-09 | | Release date: | 2014-04-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.542 Å) | | Cite: | Chromodomain Antagonists That Target the Polycomb-Group Methyllysine Reader Protein Chromobox Homolog 7 (CBX7).
J.Med.Chem., 57, 2014
|
|
3SDK
 
 | | Structure of yeast 20S open-gate proteasome with Compound 34 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(2S)-3-(3-tert-butyl-1,2,4-oxadiazol-5-yl)-1-({(2S)-1-[(4-methylbenzyl)amino]-1-oxo-4-phenylbutan-2-yl}amino)-1-oxopropan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-06-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7FBS
 
 | | structure of a channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1-[2-[(2R)-2-oxidanyl-3-(propylamino)propoxy]phenyl]-3-phenyl-propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D.J, Catterall, W.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-09-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Open-state structure and pore gating mechanism of the cardiac sodium channel.
Cell, 184, 2021
|
|
4HA3
 
 | | Structure of beta-glycosidase from Acidilobus saccharovorans in complex with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonov, A.V, Bezsudnova, E.Y, Dorovatovskii, P.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structures of beta-glycosidase from Acidilobus saccharovorans in complexes with tris and glycerol.
Dokl.Biochem.Biophys., 449
|
|
7JOV
 
 | |
