4K8Z
 
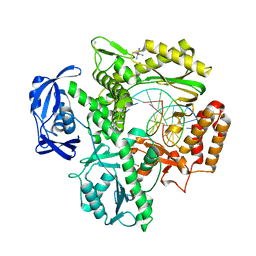 | | KOD Polymerase in binary complex with dsDNA | | Descriptor: | 1,2-ETHANEDIOL, COBALT HEXAMMINE(III), DNA (5'-D(*AP*AP*AP*TP*TP*CP*GP*CP*AP*GP*TP*TP*CP*GP*CP*G)-3'), ... | | Authors: | Bergen, K, Betz, K, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of KOD and 9N DNA Polymerases Complexed with Primer Template Duplex
Chembiochem, 14, 2013
|
|
4OPJ
 
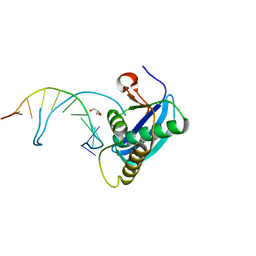 | | Bh-RNaseH:tcdA-DNA complex | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*(TCY)P*TP*TP*CP*GP*CP*G)-3', GLYCEROL, Ribonuclease H | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2014-02-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | Generating Crystallographic Models of DNA Dodecamers from Structures of RNase H:DNA Complexes.
Methods Mol.Biol., 1320
|
|
4FTL
 
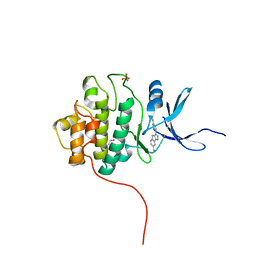 | | Crystal Structure of the CHK1 | | Descriptor: | 4'-[7-(hydroxymethyl)-2,4-dihydroindeno[1,2-c]pyrazol-3-yl]biphenyl-4-ol, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
3VRW
 
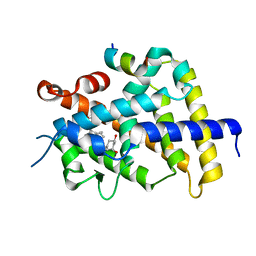 | | VDR ligand binding domain in complex with 22S-Butyl-2-methylidene-26,27-dimethyl-19,24-dinor-1alpha,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R,7E,17beta)-17-[(2R,3S)-3-butyl-5-ethyl-5-hydroxyheptan-2-yl]-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Yoshimoto, N, Inaba, Y, Itoh, T, Nakabayashi, M, Ito, N, Yamamoto, K. | | Deposit date: | 2012-04-16 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Butyl pocket formation in the vitamin d receptor strongly affects the agonistic or antagonistic behavior of ligands
J.Med.Chem., 55, 2012
|
|
6LVC
 
 | | Structure of Dimethylformamidase, dimer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5PXT
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 53) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5BVW
 
 | | Fragment-based discovery of potent and selective DDR1/2 inhibitors | | Descriptor: | Epithelial discoidin domain-containing receptor 1, IODIDE ION, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | Authors: | Murray, C, Berdini, V, Buck, I, Carr, M, Cleasby, A, Coyle, J, Curry, J, Day, J, Hearn, K, Iqbal, A, Lee, L, Martins, V, Mortenson, P, Munck, J, Page, L, Patel, S, Roomans, S, Kirsten, T, Saxty, G. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-Based Discovery of Potent and Selective DDR1/2 Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
4GBR
 
 | |
4GC5
 
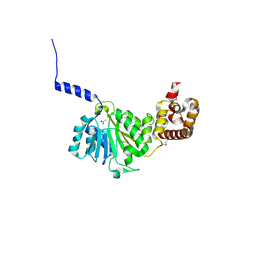 | | Crystal structure of murine TFB1M | | Descriptor: | ACETATE ION, Dimethyladenosine transferase 1, mitochondrial | | Authors: | Guja, K.E, Yakubovskaya, E, Shi, H, Mejia, E, Hambardjieva, E, Venkataraman, K, Karzai, A.W, Garcia-Diaz, M. | | Deposit date: | 2012-07-29 | | Release date: | 2013-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for S-adenosylmethionine binding and methyltransferase activity by mitochondrial transcription factor B1.
Nucleic Acids Res., 41, 2013
|
|
3O0I
 
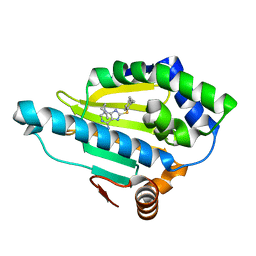 | |
4FWN
 
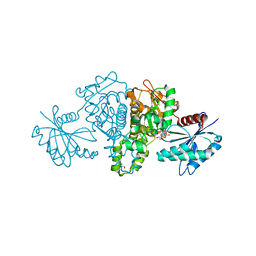 | | Crystal structure of Salmonella typhimurium propionate kinase (TdcD) in complex with Adenosine Tetraphosphate (AP4) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TETRAPHOSPHATE, Propionate kinase | | Authors: | Chittori, S, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic features of Salmonella typhimurium propionate kinase (TdcD): insights from kinetic and crystallographic studies.
Biochim.Biophys.Acta, 1834, 2013
|
|
5Q0U
 
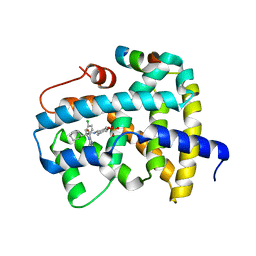 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, trans-4-({(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylacetyl}amino)cyclohexyl hydrogen sulfate | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
4CJ0
 
 | | Crystal structure of CelD in complex with affitin E12 | | Descriptor: | CALCIUM ION, E12 AFFITIN, ENDOGLUCANASE D, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
3GCV
 
 | | Human P38 MAP Kinase in Complex with RL62 | | Descriptor: | 1-{3-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2009-02-22 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors.
J.Am.Chem.Soc., 131, 2009
|
|
5Q1C
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-cyclohexyl-2-[2-(2,6-dimethoxypyridin-3-yl)-5,6-difluoro-1H-benzimidazol-1-yl]-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
6BRJ
 
 | | DDR1 bound to VX-680 | | Descriptor: | CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, Epithelial discoidin domain-containing receptor 1 | | Authors: | Georghiou, G, Seeliger, M.A. | | Deposit date: | 2017-11-30 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | What Makes a Kinase Promiscuous for Inhibitors?
Cell Chem Biol, 26, 2019
|
|
4OW1
 
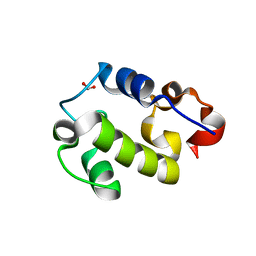 | | Crystal Structure of Resuscitation Promoting Factor C | | Descriptor: | 1,2-ETHANEDIOL, Resuscitation-promoting factor RpfC | | Authors: | Chauviac, F.X, Quay, D.H.X, Cohen-Gonsaud, M, Keep, N.H. | | Deposit date: | 2014-01-29 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The RpfC (Rv1884) atomic structure shows high structural conservation within the resuscitation-promoting factor catalytic domain.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1MND
 
 | |
1DLY
 
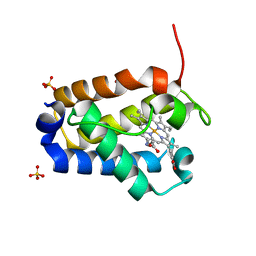 | | X-RAY CRYSTAL STRUCTURE OF HEMOGLOBIN FROM THE GREEN UNICELLULAR ALGA CHLAMYDOMONAS EUGAMETOS | | Descriptor: | 1,2-ETHANEDIOL, CYANIDE ION, HEMOGLOBIN, ... | | Authors: | Pesce, A, Couture, M, Guertin, M, Dewilde, S, Moens, L, Bolognesi, M. | | Deposit date: | 1999-12-13 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel two-over-two alpha-helical sandwich fold is characteristic of the truncated hemoglobin family.
EMBO J., 19, 2000
|
|
4CJ1
 
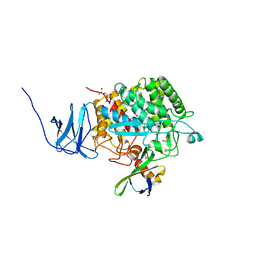 | | Crystal structure of CelD in complex with affitin H3 | | Descriptor: | CALCIUM ION, ENDOGLUCANASE D, GLYCEROL, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
6JRQ
 
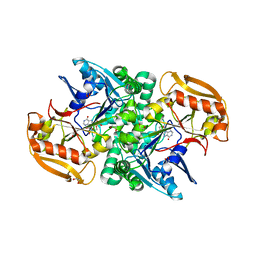 | | Crystal structure of adenylosuccinate synthetase, PurA, from Thermus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, Adenylosuccinate synthetase, INOSINIC ACID | | Authors: | Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2019-04-05 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of adenylosuccinate synthetase, PurA, from Thermus thermophilus HB8
To Be Published
|
|
3W25
 
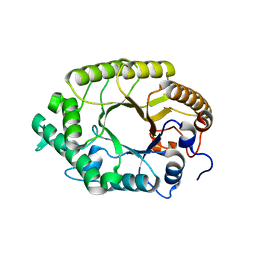 | | The high-resolution crystal structure of TsXylA, intracellular xylanase from /Thermoanaerobacterium saccharolyticum JW/SL-YS485/: the complex of the E146A mutant with xylobiose | | Descriptor: | Glycoside hydrolase family 10, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Han, X, Gao, J, Shang, N, Huang, C.-H, Ko, T.-P, Zhu, Z, Wiegel, J, Shao, W, Guo, R.-T. | | Deposit date: | 2012-11-27 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional analyses of catalytic domain of GH10 xylanase from Thermoanaerobacterium saccharolyticum JW/SL-YS485
Proteins, 81, 2013
|
|
4TMX
 
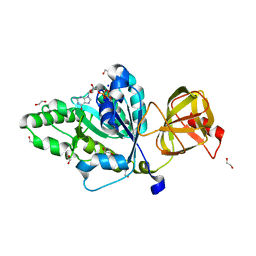 | | Translation initiation factor eIF5B (517-858) mutant D533N from C. thermophilum, bound to GTP and sodium | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kuhle, B, Ficner, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A monovalent cation acts as structural and catalytic cofactor in translational GTPases.
Embo J., 33, 2014
|
|
5PF6
 
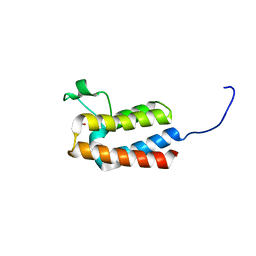 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 135) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PFT
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 157) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
