3FXZ
 
 | |
7KL5
 
 | | Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues: Phe4246 to Val4271 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-1, ... | | Authors: | Fisher, A.J, Ames, J.B, Yu, Q. | | Deposit date: | 2020-10-29 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues Phe4246-Val4271 Reveals a Fifth Calcium Binding Site.
Biochemistry, 60, 2021
|
|
3ORU
 
 | |
4DBH
 
 | | Crystal structure of Cg1458 with inhibitor | | Descriptor: | 2-HYDROXYHEPTA-2,4-DIENE-1,7-DIOATE ISOMERASE, MAGNESIUM ION, OXALATE ION | | Authors: | Ran, T.T, Wang, W.W, Xu, D.Q, Gao, Y.Y. | | Deposit date: | 2012-01-15 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of Cg1458 reveal a catalytic lid domain and a common catalytic mechanism for FAH family.
Biochem.J., 449, 2013
|
|
3G7R
 
 | | Crystal structure of SCO4454, a TetR-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Chang, C, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-10 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure and ligand specificity of SCO4454, a TetR-family transcriptional regulator from Streptomyces coelicolor
To be Published
|
|
7KIC
 
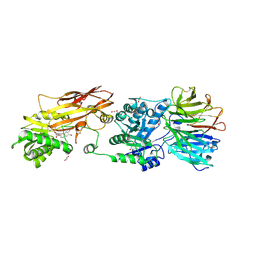 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 34 | | Descriptor: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, Methylosome protein 50, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
4DUH
 
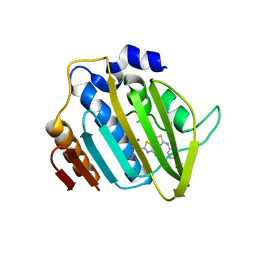 | | Crystal structure of 24 kDa domain of E. coli DNA gyrase B in complex with small molecule inhibitor | | Descriptor: | 4-{[4'-methyl-2'-(propanoylamino)-4,5'-bi-1,3-thiazol-2-yl]amino}benzoic acid, DNA gyrase subunit B | | Authors: | Brvar, M, Renko, M, Perdih, A, Solmajer, T, Turk, D. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based discovery of substituted 4,5'-bithiazoles as novel DNA gyrase inhibitors.
J.Med.Chem., 55, 2012
|
|
7L9Y
 
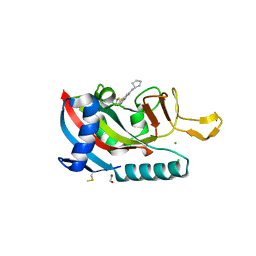 | | Human PARP14 (ARTD8), catalytic fragment in complex with RBN012042 | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopentylamino)-5-fluoro-2-{[(piperidin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, CHLORIDE ION, ... | | Authors: | Dorsey, B.W, Swinger, K.K, Schenkel, L.B, Church, W.D, Perl, N.R, Vasbinder, M.M, Wigle, T.J, Kuntz, K.W. | | Deposit date: | 2021-01-05 | | Release date: | 2021-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeted Degradation of PARP14 Using a Heterobifunctional Small Molecule.
Chembiochem, 22, 2021
|
|
3ZKJ
 
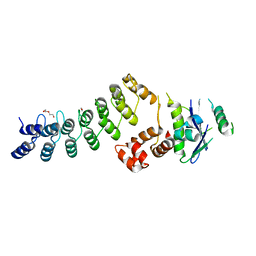 | | Crystal Structure of Ankyrin Repeat and Socs Box-Containing Protein 9 (Asb9) in Complex with Elonginb and Elonginc | | Descriptor: | 1,2-ETHANEDIOL, ANKYRIN REPEAT AND SOCS BOX PROTEIN 9, CHLORIDE ION, ... | | Authors: | Muniz, J.R.C, Guo, K, Zhang, Y, Ayinampudi, V, Savitsky, P, Keates, T, Filippakopoulos, P, Vollmar, M, Yue, W.W, Krojer, T, Ugochukwu, E, von Delft, F, Knapp, S, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2013-01-23 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
1S8L
 
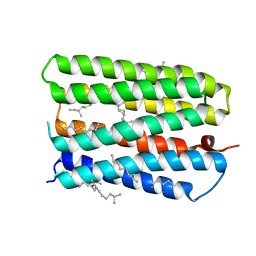 | | Anion-free form of the D85S mutant of bacteriorhodopsin from crystals grown in the presence of halide | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin precursor, RETINAL | | Authors: | Facciotti, M.T, Cheung, V.S, Lunde, C.S, Rouhani, S, Baliga, N.S, Glaeser, R.M. | | Deposit date: | 2004-02-02 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Specificity of anion binding in the substrate pocket of bacteriorhodopsin.
Biochemistry, 43, 2004
|
|
3L0B
 
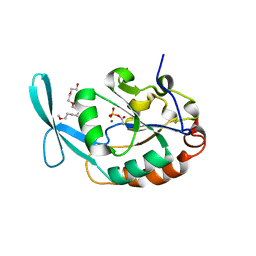 | | Crystal structure of SCP1 phosphatase D206A mutant phosphoryl-intermediate | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2009-12-09 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1.
Protein Sci., 19, 2010
|
|
3L0Y
 
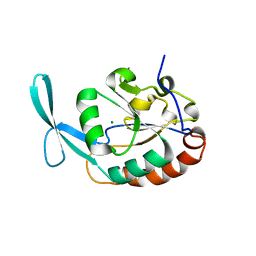 | | Crystal structure OF SCP1 phosphatase D98A mutant | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION | | Authors: | Zhang, M, Zhang, Y. | | Deposit date: | 2009-12-10 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1.
Protein Sci., 19, 2010
|
|
2Z72
 
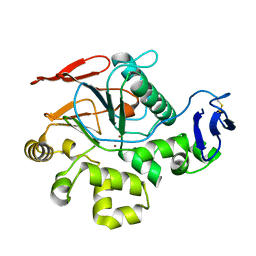 | | New Structure Of Cold-Active Protein Tyrosine Phosphatase At 1.1 Angstrom | | Descriptor: | Protein-tyrosine-phosphatase, ZINC ION | | Authors: | Tsuruta, H, Mikami, B, Yamamoto, C, Yamagata, H. | | Deposit date: | 2007-08-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The role of group bulkiness in the catalytic activity of psychrophile cold-active protein tyrosine phosphatase
Febs J., 275, 2008
|
|
3ZZF
 
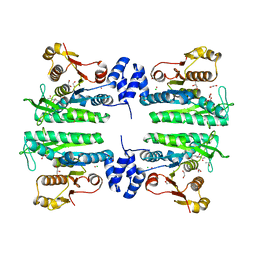 | | Crystal structure of the amino acid kinase domain from Saccharomyces cerevisiae acetylglutamate kinase complexed with its substrate N- acetylglutamate | | Descriptor: | 1,2-ETHANEDIOL, ACETYLGLUTAMATE KINASE, CHLORIDE ION, ... | | Authors: | de Cima, S, Gil-Ortiz, F, Crabeel, M, Fita, I, Rubio, V. | | Deposit date: | 2011-09-01 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insight on an Arginine Synthesis Metabolon from the Tetrameric Structure of Yeast Acetylglutamate Kinase
Plos One, 7, 2012
|
|
4M8R
 
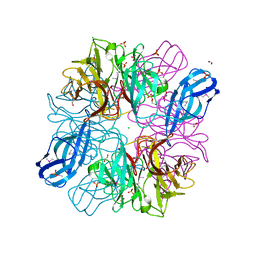 | |
3G3V
 
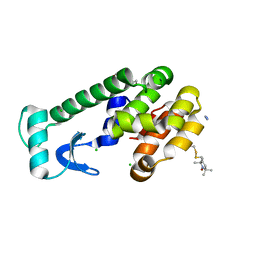 | | Crystal structure of spin labeled T4 Lysozyme (V131R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
4DEU
 
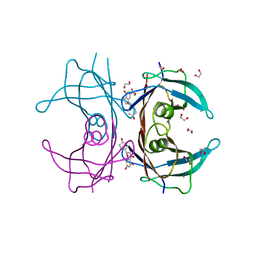 | |
1HSX
 
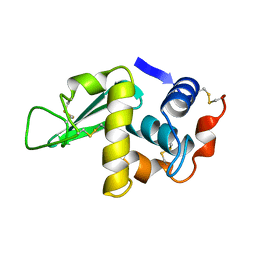 | |
1WVX
 
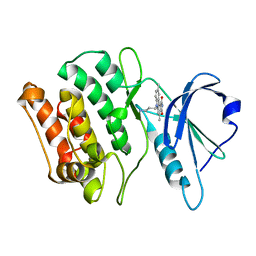 | | Crystal structures of kinase domain of DAP kinase in complex with small molecular inhibitors | | Descriptor: | 6-(3-AMINOPROPYL)-4,9-DIMETHYLPYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, Death-associated protein kinase 1 | | Authors: | Ueda, Y, Ogata, H, Yamakawa, A, Higuchi, Y. | | Deposit date: | 2004-12-27 | | Release date: | 2006-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complex structure of kinase domain of DAP kinase with BDB402
To be Published
|
|
3G64
 
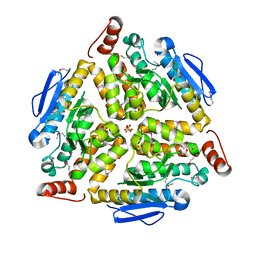 | | Crystal structure of putative enoyl-CoA hydratase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Putative enoyl-CoA hydratase, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Putative Enoyl-CoA Hydratase from Streptomyces coelicolor A3(2)
To be Published
|
|
5OUO
 
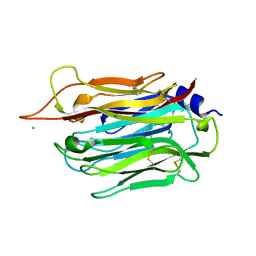 | | Structure of TgPLP1 APCbeta domain | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Perforin-like protein 1 | | Authors: | Ni, T, Gilbert, R.J.C. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structures of monomeric and oligomeric forms of theToxoplasma gondiiperforin-like protein 1.
Sci Adv, 4, 2018
|
|
1HOV
 
 | | SOLUTION STRUCTURE OF A CATALYTIC DOMAIN OF MMP-2 COMPLEXED WITH SC-74020 | | Descriptor: | CALCIUM ION, MATRIX METALLOPROTEINASE-2, N-{4-[(1-HYDROXYCARBAMOYL-2-METHYL-PROPYL)-(2-MORPHOLIN-4-YL-ETHYL)-SULFAMOYL]-4-PENTYL-BENZAMIDE, ... | | Authors: | Feng, Y, Likos, J.J, Zhu, L, Woodward, H, Munie, G, McDonald, J.J, Stevens, A.M, Howard, C.P, De Crescenzo, G.A, Welsch, D, Shieh, H.-S, Stallings, W.C. | | Deposit date: | 2000-12-11 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the catalytic domain of matrix metalloproteinase-2 complexed with a hydroxamic acid inhibitor
Biochim.Biophys.Acta, 1598, 2002
|
|
4DJV
 
 | | Structure of BACE Bound to 2-imino-5-(3'-methoxy-[1,1'-biphenyl]-3-yl)-3-methyl-5-phenylimidazolidin-4-one | | Descriptor: | (2E,5R)-2-imino-5-(3'-methoxybiphenyl-3-yl)-3-methyl-5-phenylimidazolidin-4-one, Beta-secretase 1, L(+)-TARTARIC ACID | | Authors: | Strickland, C, Cumming, J. | | Deposit date: | 2012-02-02 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure based design of iminohydantoin BACE1 inhibitors: Identification of an orally available, centrally active BACE1 inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3ODR
 
 | |
3G3W
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
