1MZE
 
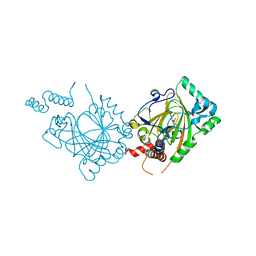 | | Human Factor Inhibiting HIF (FIH1) | | Descriptor: | D(-)-TARTARIC ACID, FE (II) ION, factor inhibiting HIF1 | | Authors: | Dann III, C.E, Bruick, R.K, Deisenhofer, J. | | Deposit date: | 2002-10-07 | | Release date: | 2002-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Factor-Inhibiting Hypoxia-Inducible Factor 1: An Asparaginyl Hydroxylase Involved in the Hypoxic Response Pathway
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5QE1
 
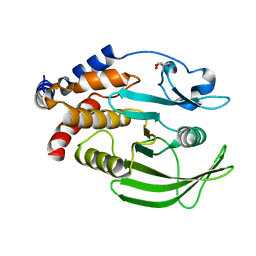 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000645a | | Descriptor: | (2,3-dihydro-1,2,3-benzothiadiazol-5-yl)(morpholin-4-yl)methanone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5YAV
 
 | | Fragment-based Drug Discovery of inhibitors to block PDEdelta-RAS protein-protein interaction | | Descriptor: | 1-phenyl-5-propan-2-ylsulfanyl-1,2,3,4-tetrazole, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Bing, X, Yanlian, L, Danyan, C. | | Deposit date: | 2017-09-01 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Fragment-based Drug Discovery of inhibitors to block PDEdelta-RAS protein-protein interaction
To Be Published
|
|
5QEY
 
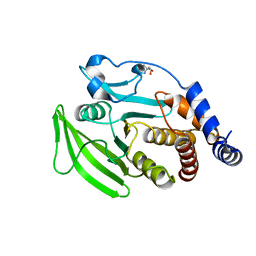 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000708a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-[5-(methylsulfanyl)-1,3,4-thiadiazol-2-yl]furan-2-carboxamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5YB6
 
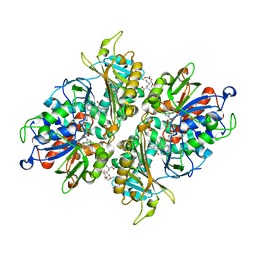 | | L-Amino acid oxidase/monooxygenase from Pseudomonas sp. AIU 813 - L-lysine complex | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase/monooxygenase, ... | | Authors: | Im, D, Matsui, D, Arakawa, T, Isobe, K, Asano, Y, Fushinobu, S. | | Deposit date: | 2017-09-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand complex structures of l-amino acid oxidase/monooxygenase from
FEBS Open Bio, 8, 2018
|
|
5QFP
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000293a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QG9
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOOA000595a | | Descriptor: | (1S,3S,4R)-3-[(1S)-1-hydroxypropyl]-2-(methylsulfonyl)-2-azabicyclo[2.2.2]octan-4-ol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.671 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
3JAB
 
 | | Domain organization and conformational plasticity of the G protein effector, PDE6 | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, GafA domain of cone phosphodiesterase 6C, GafB domain of phosphodiesterase 2A, ... | | Authors: | Zhang, Z, He, F, Constantine, R, Baker, M.L, Baehr, W, Schmid, M.F, Wensel, T.G, Agosto, M.A. | | Deposit date: | 2015-05-26 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Domain Organization and Conformational Plasticity of the G Protein Effector, PDE6.
J.Biol.Chem., 290, 2015
|
|
5TR6
 
 | | Discovery of TAK-659, an Orally Available Investigational Inhibitor of Spleen Tyrosine Kinase (SYK) | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(1R,2S)-2-aminocyclohexyl]amino}-7-fluoro-4-(1-methyl-1H-pyrazol-4-yl)-1,2-dihydro-3H-pyrrolo[3,4-c]pyridin-3-one, Tyrosine-protein kinase SYK | | Authors: | Yano, J, Jennings, A, Lam, B, Hoffman, I.D. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of TAK-659 an orally available investigational inhibitor of Spleen Tyrosine Kinase (SYK).
Bioorg. Med. Chem. Lett., 26, 2016
|
|
7JTV
 
 | |
5DCL
 
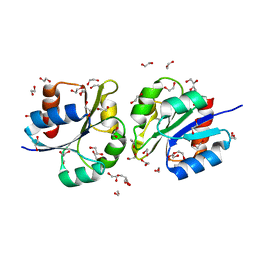 | | Structure of a lantibiotic response regulator: N terminal domain of the nisin resistance regulator NsrR | | Descriptor: | 1,2-ETHANEDIOL, PhoB family transcriptional regulator | | Authors: | Khosa, S, Kleinschrodt, D, Hoeppner, A, Smits, S.H. | | Deposit date: | 2015-08-24 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structure of the Response Regulator NsrR from Streptococcus agalactiae, Which Is Involved in Lantibiotic Resistance.
Plos One, 11, 2016
|
|
4BZC
 
 | | Crystal structure of the tetrameric dGTP-bound wild type SAMHD1 catalytic core | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), DEOXYNUCLEOSIDE TRIPHOSPHATE TRIPHOSPHOHYDROLASE SAMHD1, MAGNESIUM ION, ... | | Authors: | Ji, X, Yang, H, Wu, Y, Yan, J, Mehrens, J, DeLucia, M, Hao, C, Gronenborn, A.M, Skowronski, J, Ahn, J, Xiong, Y. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Mechanism of Allosteric Activation of Samhd1 by Dgtp
Nat.Struct.Mol.Biol., 20, 2013
|
|
4ZRM
 
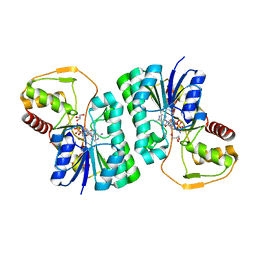 | |
2VZC
 
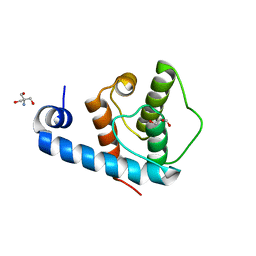 | | Crystal structure of the C-terminal calponin homology domain of alpha parvin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Analysis of the Interactions between Paxillin Ld Motifs and Alpha-Parvin
Structure, 16, 2008
|
|
8F1S
 
 | | A benzimidazole (DB1476) sequence-specific recognition of 5'-CGCAAAAAAGCG-3' in A-orientation | | Descriptor: | 4,4'-(1H-benzimidazole-2,6-diyl)di(benzene-1-carboximidamide), DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3'), ... | | Authors: | Ogbonna, E.N, Wilson, W.D. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure Characterization of the Selective Recognition of AT Base Pair Sequences.
Acs Bio Med Chem Au, 3, 2023
|
|
7ELC
 
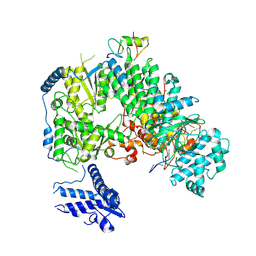 | | Structure of monomeric complex of MACV L bound to Z and 3'-vRNA | | Descriptor: | 3'-vRNA promoter, MANGANESE (II) ION, RING finger protein Z, ... | | Authors: | Xu, X, Peng, R, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
3JS3
 
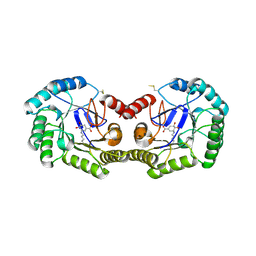 | | Crystal structure of type I 3-dehydroquinate dehydratase (aroD) from Clostridium difficile with covalent reaction intermediate | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, 3-dehydroquinate dehydratase | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-09 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.
J.Biol.Chem., 286, 2011
|
|
5TVY
 
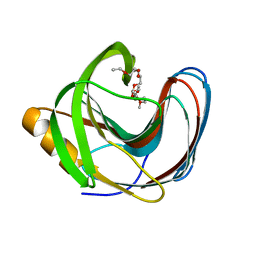 | | Computationally Designed Fentanyl Binder - Fen49 | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Endo-1,4-beta-xylanase A | | Authors: | Bick, M.J, Greisen, P.J, Morey, K.J, Antunes, M.S, La, D, Sankaran, B, Reymond, L, Johnsson, K, Medford, J.I, Baker, D. | | Deposit date: | 2016-11-10 | | Release date: | 2017-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of environmental sensors for the potent opioid fentanyl.
Elife, 6, 2017
|
|
3QQG
 
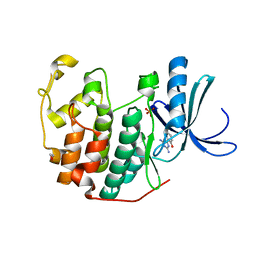 | | CDK2 in complex with inhibitor L2-5 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-(4,6-diamino-1,3,5-triazin-2-yl)phenol, Cyclin-dependent kinase 2, ... | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-15 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3DMC
 
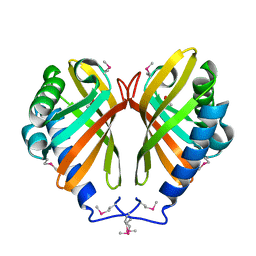 | |
4PF7
 
 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | (2S)-2-amino-N-{(1S)-1-cyclohexyl-2-[(4-methylphenyl)amino]-2-oxoethyl}-4-(methylselanyl)butanamide, Insulin-degrading enzyme, ZINC ION | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
6MGH
 
 | | X-ray structure of monomeric near-infrared fluorescent protein miRFP670nano | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(3~{S},4~{R})-3-ethenyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Pletnev, S. | | Deposit date: | 2018-09-13 | | Release date: | 2018-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing.
Nat Commun, 10, 2019
|
|
5TY9
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-(2,4-dimethoxyphenyl)-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-18 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5U0F
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-[(2,4-dimethoxyphenyl)methyl]-2-sulfanylidene-1,3-thiazolidin-4-one, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
7ELB
 
 | | Structure of Machupo virus L polymerase in complex with Z protein (dimeric form) | | Descriptor: | MANGANESE (II) ION, RING finger protein Z, RNA-directed RNA polymerase L, ... | | Authors: | Peng, R, Xu, X, Peng, Q, Shi, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of Lassa and Machupo virus polymerases complexed with cognate regulatory Z proteins identify targets for antivirals.
Nat Microbiol, 6, 2021
|
|
