5ADT
 
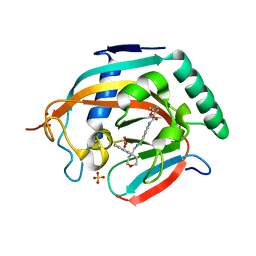 | | Crystal structure of human tankyrase 2 in complex with OD73 | | Descriptor: | BICARBONATE ION, N-[3-chloranyl-4-[[4-(4-methoxyphenyl)oxan-4-yl]methylcarbamoyl]phenyl]-2-methyl-1,3-oxazole-5-carboxamide, SULFATE ION, ... | | Authors: | Haikarainen, T, Lehtio, L. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development and Structural Analysis of Adenosine Site Binding Tankyrase Inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
1VFO
 
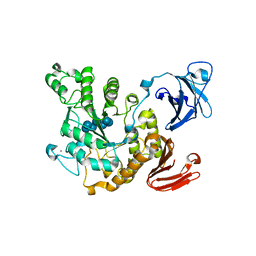 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/beta-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclic alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-16 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
5VWA
 
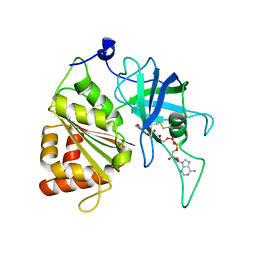 | | Y316F mutant of corn root ferredoxin:NADP+ reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase, MAGNESIUM ION | | Authors: | Kean, K.M, Carpenter, R.A, Hall, A.R, Karplus, P.A. | | Deposit date: | 2017-05-21 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution studies of hydride transfer in the ferredoxin:NADP(+) reductase superfamily.
FEBS J., 284, 2017
|
|
3J1V
 
 | |
6UCX
 
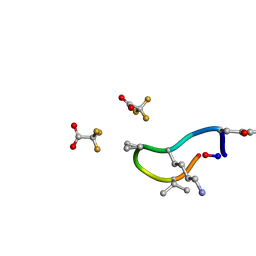 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
1A4V
 
 | | ALPHA-LACTALBUMIN | | Descriptor: | ALPHA-LACTALBUMIN, CALCIUM ION | | Authors: | Chandra, N, Acharya, K.R. | | Deposit date: | 1998-02-05 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evidence for the presence of a secondary calcium binding site in human alpha-lactalbumin.
Biochemistry, 37, 1998
|
|
3ZUN
 
 | | pVHL54-213-EloB-EloC complex_(2S,4R)-4-hydroxy-1-(2-(3-methylisoxazol- 5-yl)acetyl)-N-(4-nitrobenzyl)pyrrolidine-2-carboxamide bound | | Descriptor: | (4R)-N-[4-(DIHYDROXYAMINO)BENZYL]-4-HYDROXY-1-[(3-METHYLISOXAZOL-5-YL)ACETYL]-L-PROLINAMIDE, GLYCEROL, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Van Molle, I, Buckley, D, Crews, C.M, Ciulli, A. | | Deposit date: | 2011-07-19 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Elongin-B, Elongin-C, Von Hippel-Lindau Disease Tumor Suppressor Complex
To be Published
|
|
6UDZ
 
 | | S2 symmetric peptide design number 4 crystal form 1, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 1, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
159L
 
 | | CONTROL OF ENZYME ACTIVITY BY AN ENGINEERED DISULFIDE BOND | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Blaber, M, Matthews, B.W. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alanine scanning mutagenesis of the alpha-helix 115-123 of phage T4 lysozyme: effects on structure, stability and the binding of solvent.
J.Mol.Biol., 246, 1995
|
|
2YPJ
 
 | | Non-catalytic carbohydrate binding module CBM65B | | Descriptor: | ENDOGLUCANASE CEL5A, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Temple, M.J, Rogowski, A, Basle, A, Xue, J, Knox, J.P, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan
J.Biol.Chem., 288, 2013
|
|
5AIZ
 
 | | The PIAS-like coactivator Zmiz1 is a direct and selective cofactor of Notch1 in T-cell development and leukemia | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Cho, H.J, Murai, M, Chiang, M, Cierpicki, T. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Pias-Like Coactivator Zmiz1 is a Direct and Selective Cofactor of Notch1 in T-Cell Development and Leukemia
Immunity, 43, 2015
|
|
7F35
 
 | | Crystal structure of anti S-gatifloxacin antibody Fab fragment in complex with S-gatifloxacin | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-6-fluoro-8-methoxy-7-[(3S)-3-methylpiperazin-1-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, Antibody Fab fragment heavy chain, ... | | Authors: | Wang, L.T, Jiao, W.Y, Shen, X, Lei, H.T. | | Deposit date: | 2021-06-15 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational adaptability determining antibody recognition to distomer: structure analysis of enantioselective antibody against chiral drug gatifloxacin
Rsc Adv, 11, 2021
|
|
5EDD
 
 | | Crystal structure of Mycobacterium tuberculosis dUTPase R140K, H145W mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Nagy, G.N, Leveles, I, Lopata, A, Harmat, V, Toth, J, Vertessy, G.B. | | Deposit date: | 2015-10-21 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Characterization of Arginine Fingers: Identification of an Arginine Finger for the Pyrophosphatase dUTPases.
J. Am. Chem. Soc., 138, 2016
|
|
7EWA
 
 | | GDP-bound KRAS G12D in complex with TH-Z827 | | Descriptor: | 4-[(1~{R},5~{S})-3,8-diazabicyclo[3.2.1]octan-8-yl]-7-(8-methylnaphthalen-1-yl)-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
5A44
 
 | | Structure of Bacteriorhodopsin obtained from 20um crystals by multi crystal data collection | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, BACTERIORHODOPSIN, HEPTANE, ... | | Authors: | Zander, U, Bourenkov, G, Popov, A.N, de Sanctis, D, McCarthy, A.A, Svensson, O, Round, E.S, Gordeliy, V.I, Mueller-Dieckmann, C, Leonard, G.A. | | Deposit date: | 2015-06-05 | | Release date: | 2015-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Meshandcollect: An Automated Multi-Crystal Data-Collection Workflow for Synchrotron Macromolecular Crystallography Beamlines.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7KDR
 
 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate inhibitor HP-75 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-{[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]sulfonyl}-5'-deoxyadenosine, ... | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.488 Å) | | Cite: | Bisubstrate inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Transition state analogs for high affinity binding.
Bioorg.Med.Chem., 29, 2021
|
|
5NZ3
 
 | | The structure of the thermobifida fusca guanidine III riboswitch with methylguanidine | | Descriptor: | 1-METHYLGUANIDINE, MAGNESIUM ION, RNA (41-MER), ... | | Authors: | Huang, L, Wang, J, Lilley, D.M.J. | | Deposit date: | 2017-05-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Structure of the Guanidine III Riboswitch.
Cell Chem Biol, 24, 2017
|
|
5DT7
 
 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group C2221 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
169L
 
 | |
7FB7
 
 | | Crystal structure of human UHRF1 TTD in complex with 5-amino-2,4-dimethylpyridine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-amino-2,4-dimethylpyridine, DIMETHYL SULFOXIDE, ... | | Authors: | Kori, S, Arita, K, Yoshimi, S. | | Deposit date: | 2021-07-08 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1.
Bioorg.Med.Chem., 52, 2021
|
|
6IWH
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*05104 complexed with C14-GGGI lipopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, C14-GGGI lipopeptide, ... | | Authors: | Yamamoto, Y, Morita, D, Sugita, M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification and Structure of an MHC Class I-Encoded Protein with the Potential to PresentN-Myristoylated 4-mer Peptides to T Cells.
J Immunol., 202, 2019
|
|
7KDZ
 
 | |
5PON
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10980a | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(pyridin-2-yl)piperazin-1-yl]ethan-1-one, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.519 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3ZZK
 
 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, GLYCEROL, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
4CXO
 
 | | bifunctional endonuclease in complex with ssDNA | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ENDONUCLEASE 2, SULFATE ION, ... | | Authors: | Yu, T.-F, Maestre-Reyna, M, Ko, C.-Y, Ko, T.-P, Sun, Y.-J, Lin, T.-Y, Shaw, J.-F, Wang, A.H.-J. | | Deposit date: | 2014-04-08 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights of the Ssdna Binding Site in the Multifunctional Endonuclease Atbfn2 from Arabidopsis Thaliana.
Plos One, 9, 2014
|
|
