7SGR
 
 | | Structure of hemolysin A secretion system HlyB/D complex | | Descriptor: | Alpha-hemolysin translocation ATP-binding protein HlyB, Membrane fusion protein (MFP) family protein,Hemolysin secretion protein D, chromosomal, ... | | Authors: | Zhao, H, Chen, J. | | Deposit date: | 2021-10-07 | | Release date: | 2022-09-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The hemolysin A secretion system is a multi-engine pump containing three ABC transporters.
Cell, 185, 2022
|
|
7SH1
 
 | | Class II UvrA protein - Ecm16 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Excinuclease ABC subunit UvrA, ... | | Authors: | Grade, P, Erlandson, A, Ullah, A, Mathews, I.I, Chen, X, Kim, C.-Y, Mera, P.E. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional analyses of the echinomycin resistance conferring protein Ecm16 from Streptomyces lasalocidi.
Sci Rep, 13, 2023
|
|
7PX4
 
 | | Crystal structure of the adenosine A2A receptor (A2A-PSB1-bRIL) in complex with preladenant conjugate PSB-2113 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Claff, T, Klapschinski, T.A, Tiruttani Subhramanyam, U.K, Vaassen, V.J, Schlegel, J.G, Vielmuth, C, Voss, J.H, Labahn, J, Muller, C.E. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Single Stabilizing Point Mutation Enables High-Resolution Co-Crystal Structures of the Adenosine A 2A Receptor with Preladenant Conjugates.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7SH2
 
 | | Structure of the yeast Rad24-RFC loader bound to DNA and the open 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SGZ
 
 | | Structure of the yeast Rad24-RFC loader bound to DNA and the closed 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7VM7
 
 | | Crystal structure of inactive uPA in complex with nafamostat | | Descriptor: | (6-carbamimidoylnaphthalen-2-yl) 4-carbamimidamidobenzoate, Urokinase-type plasminogen activator chain B | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7VM4
 
 | | Crystal structure of uPA in complex with nafamostat | | Descriptor: | 4-carbamimidamidobenzoic acid, TRIETHYLENE GLYCOL, Urokinase-type plasminogen activator | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7VM5
 
 | |
7VM6
 
 | | Crystal structure of uPA in complex with 6-amidino-2-naphthol | | Descriptor: | 6-oxidanylnaphthalene-2-carboximidamide, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L.G, Huang, M.D. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural study of the uPA-nafamostat complex reveals a covalent inhibitory mechanism of nafamostat.
Biophys.J., 121, 2022
|
|
7PWD
 
 | | Structure of an inhibited GRK2-G-beta and G-gamma complex | | Descriptor: | 4-chloranyl-N-[2-(4-chlorophenyl)ethyl]thieno[2,3-c]pyridine-2-carboxamide, Beta-adrenergic receptor kinase 1, CHLORIDE ION, ... | | Authors: | Faucher, N, Tauchert, M.J, Konz Makino, D.L, Vuillard, L.M. | | Deposit date: | 2021-10-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis of thieno[2,3-c]pyridine derived GRK2 inhibitors
Monatsh Chem, 2022
|
|
7VM2
 
 | |
7VLT
 
 | | Structure of SUR2B in complex with Mg-ATP/ADP and levcromakalim | | Descriptor: | (3S,4R)-2,2-dimethyl-3-oxidanyl-4-(2-oxidanylidenepyrrolidin-1-yl)-3,4-dihydrochromene-6-carbonitrile, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLR
 
 | | Structure of SUR2B in complex with Mg-ATP/ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLU
 
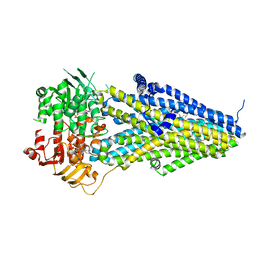 | | Structure of SUR2A in complex with Mg-ATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7VLS
 
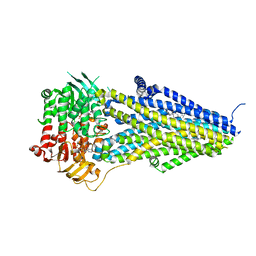 | | Structure of SUR2B in complex with MgATP/ADP and P1075 | | Descriptor: | 1-cyano-2-(2-methylbutan-2-yl)-3-pyridin-3-yl-guanidine, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Ding, D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural identification of vasodilator binding sites on the SUR2 subunit.
Nat Commun, 13, 2022
|
|
7PVR
 
 | |
7PVQ
 
 | |
7PVX
 
 | |
7PW2
 
 | |
7PVS
 
 | |
7PVV
 
 | |
7PW0
 
 | |
7PVT
 
 | |
7PVW
 
 | |
7PVY
 
 | |
