5Y02
 
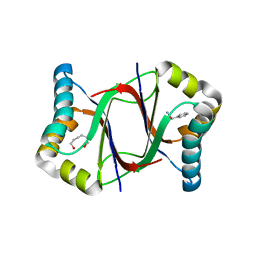 | | C-terminal peptide depleted mutant of hydroxynitrile lyase from Passiflora edulis (PeHNL) bound with (R)-mandelonitrile | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, HEXANE-1,6-DIOL, Hydroxynitrile lyase, ... | | Authors: | Motojima, F, Nuylert, A, Asano, Y. | | Deposit date: | 2017-07-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure and catalytic mechanism of hydroxynitrile lyase from passion fruit, Passiflora edulis
FEBS J., 285, 2018
|
|
1NCB
 
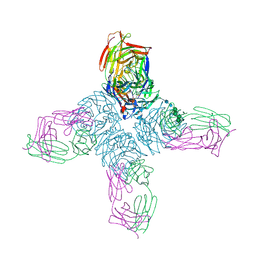 | | CRYSTAL STRUCTURES OF TWO MUTANT NEURAMINIDASE-ANTIBODY COMPLEXES WITH AMINO ACID SUBSTITUTIONS IN THE INTERFACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1992-01-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of two mutant neuraminidase-antibody complexes with amino acid substitutions in the interface.
J.Mol.Biol., 227, 1992
|
|
7AJ9
 
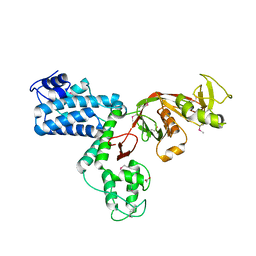 | |
7DNC
 
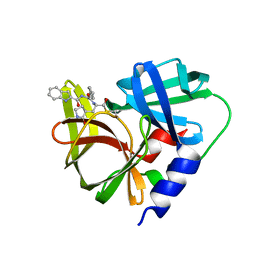 | | Crystal structure of EV71 3C proteinase in complex with a novel inhibitor | | Descriptor: | 3C protease, ~{N}-[(2~{S})-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-05 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Peptidomimetic Aldehydes as Broad-Spectrum Inhibitors against Enterovirus and SARS-CoV-2.
J.Med.Chem., 65, 2022
|
|
2B8T
 
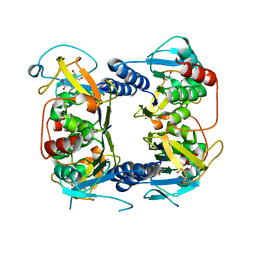 | | Crystal structure of Thymidine Kinase from U.urealyticum in complex with thymidine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, THYMIDINE, Thymidine kinase, ... | | Authors: | Kosinska, U, Carnrot, C, Eriksson, S, Wang, L, Eklund, H. | | Deposit date: | 2005-10-10 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the substrate complex of thymidine kinase from Ureaplasma urealyticum and investigations of possible drug targets for the enzyme
FEBS Lett., 272, 2005
|
|
4D3W
 
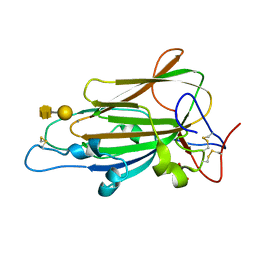 | | Crystal Structure of Epithelial Adhesin 1 A domain (Epa1A) from Candida glabrata in complex with the T-antigen (Galb1-3GalNAc) | | Descriptor: | CALCIUM ION, EPITHELIAL ADHESIN 1, GLYCEROL, ... | | Authors: | Kock, M, Maestre-Reyna, M, Diderrich, R, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2014-10-24 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Hotspots Determine Functional Diversity of the Candida Glabrata Epithelial Adhesin Family
J.Biol.Chem., 290, 2015
|
|
2B25
 
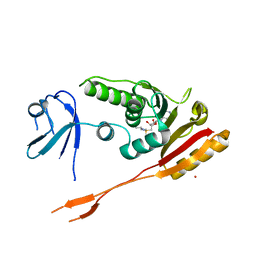 | | Human putative tRNA(1-methyladenosine)methyltransferase | | Descriptor: | S-ADENOSYLMETHIONINE, UNKNOWN ATOM OR ION, hypothetical protein | | Authors: | Tempel, W, Wu, H, Dong, A, Zeng, H, Loppnau, P, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-16 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human putative tRNA(1-methyladenosine)methyltransferase
To be Published
|
|
6M5V
 
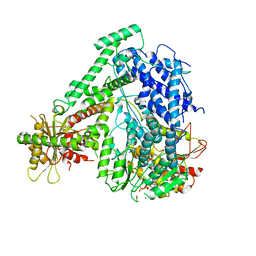 | | The coordinate of the hexameric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
4R37
 
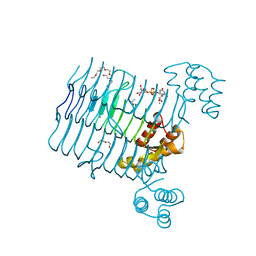 | | Crystal structure analysis of LpxA, a UDP-N-acetylglucosamine acyltransferase from Bacteroides fragilis 9343 with UDP-GlcNAc | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETATE ION, ... | | Authors: | Fisher, A.J, Chen, X, Ngo, A. | | Deposit date: | 2014-08-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Bacteroides fragilis uridine 5'-diphosphate-N-acetylglucosamine (UDP-GlcNAc) acyltransferase (BfLpxA).
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4ZY5
 
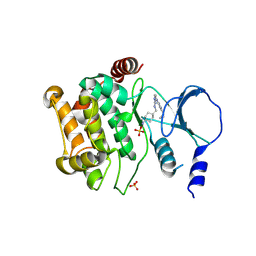 | |
7E36
 
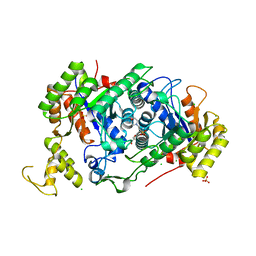 | | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkanesulfonate monooxygenase SsuD/methylene tetrahydromethanopterin reductase-like flavin-dependent oxidoreductase (Luciferase family), ... | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis.
Nat Commun, 12, 2021
|
|
6BQL
 
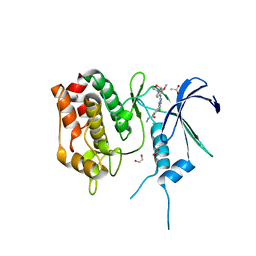 | | Crystal Structure of the Human CAMKK2B in complex with TAE-226 | | Descriptor: | 1,2-ETHANEDIOL, 2-({5-CHLORO-2-[(2-METHOXY-4-MORPHOLIN-4-YLPHENYL)AMINO]PYRIMIDIN-4-YL}AMINO)-N-METHYLBENZAMIDE, ACETATE ION, ... | | Authors: | Counago, R.M, de Souza, G.P, dos Reis, C.V, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human CAMKK2B in complex with TAE-226
To Be Published
|
|
6LXO
 
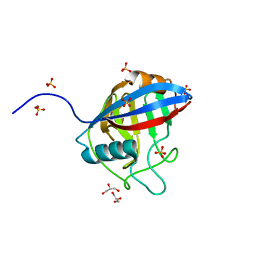 | | TvCyP2 in apo form 1 | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase, SULFATE ION | | Authors: | Aryal, S, Chen, C, Hsu, C.H. | | Deposit date: | 2020-02-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | N-Terminal Segment of TvCyP2 Cyclophilin fromTrichomonas vaginalisIs Involved in Self-Association, Membrane Interaction, and Subcellular Localization.
Biomolecules, 10, 2020
|
|
9J5W
 
 | | Cryo-EM Structure of URAT1 in Complex with Benzbromarone | | Descriptor: | Solute carrier family 22 member 12, [3,5-bis(bromanyl)-4-oxidanyl-phenyl]-(2-ethyl-1-benzofuran-3-yl)methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2024-08-13 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis for Inhibition of Urate Reabsorption in URAT1.
Jacs Au, 5, 2025
|
|
4QVL
 
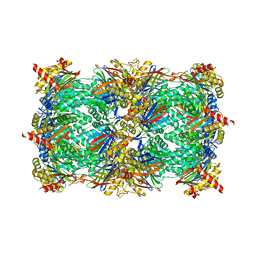 | | yCP in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5AG5
 
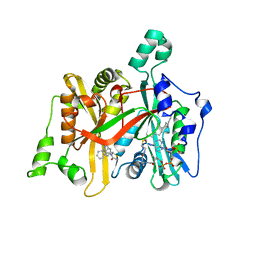 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A THIAZOLIDINONE LIGAND | | Descriptor: | (2R)-3-benzyl-2-(1H-indazol-5-yl)-1,3-thiazolidin-4-one, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Spinks, D, Smith, V.C, Thompson, S, Smith, A, Torrie, L.S, McElroy, S.P, Brand, S, Brenk, R, Frearson, J.A, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2015-01-29 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of Small-Molecule Trypanosoma Brucei N-Myristoyltransferase Inhibitors: Discovery and Optimisation of a Novel Binding Mode.
Chemmedchem, 10, 2015
|
|
5AIB
 
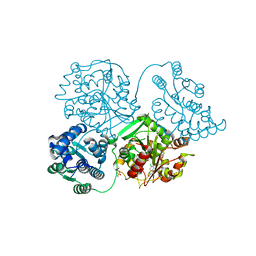 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 6-chloro-2H-1,4-benzoxazin-3(4H)-one, BIFUNCTIONAL EPOXIDE HYDROLASE 2, SULFATE ION | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-02-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
6BVT
 
 | | Importin alpha 1 in cargo free state | | Descriptor: | Importin subunit alpha-1 | | Authors: | Smith, K.M, Tsimablyuk, S, Edwards, M.R, Aragao, D, Cross, E.M, Basler, C.F, Forwood, J.K. | | Deposit date: | 2017-12-13 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses.
Nat Commun, 9, 2018
|
|
2B33
 
 | |
2B0V
 
 | | NUDIX hydrolase from Nitrosomonas europaea. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NUDIX hydrolase, ... | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
6UQN
 
 | |
6UXO
 
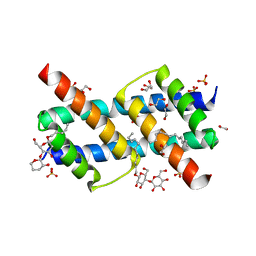 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with DDM | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bcl-2 homologous antagonist/killer, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5SVL
 
 | | Crystal structure of the ATP-gated human P2X3 ion channel in the ATP-bound, closed (desensitized) state | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mansoor, S.E, Lu, W, Oosterheert, W, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2016-08-06 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structures define human P2X3 receptor gating cycle and antagonist action.
Nature, 538, 2016
|
|
6UQV
 
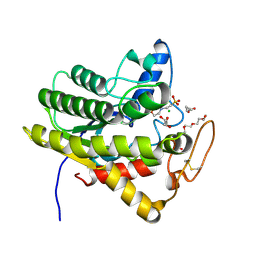 | | Crystal structure of ChoE, a bacterial acetylcholinesterase from Pseudomonas aeruginosa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ChoE, ... | | Authors: | Shi, R, Pham, V.D, To, T.A. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-13 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into the putative bacterial acetylcholinesterase ChoE and its substrate inhibition mechanism.
J.Biol.Chem., 295, 2020
|
|
5Y0T
 
 | |
