7CMF
 
 | | Crystal structure of human P-cadherin REC12 (monomer) in complex with 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine (inhibitor) | | Descriptor: | 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3 | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of cadherin dimerization by chemical fragments as a trigger to inhibit cell adhesion
Commun Biol, 4, 2021
|
|
4OBA
 
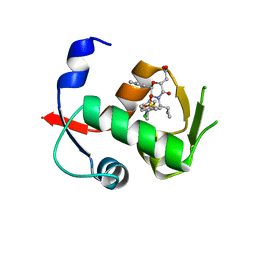 | | Co-crystal structure of MDM2 with Inhibitor Compound 4 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, [(2R,5R,6R)-4-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(3-chlorophenyl)-5-(4-chlorophenyl)-3-oxomorpholin-2-yl]acetic acid | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Selective and Potent Morpholinone Inhibitors of the MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 57, 2014
|
|
5DX6
 
 | |
6LEI
 
 | |
4F8C
 
 | |
6AYO
 
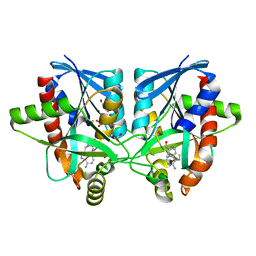 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 5'-deoxy-5'-Propyl-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-propylpyrrolidin-3-ol, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
7HCA
 
 | |
8AYE
 
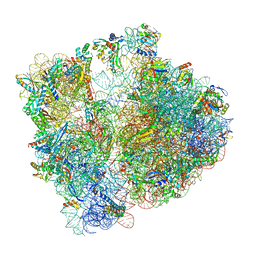 | | E. coli 70S ribosome bound to thermorubin and fMet-tRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Sanyal, S, Parajuli, N.P, Emmerich, A.G. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-01 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | Antibiotic thermorubin tethers ribosomal subunits and impedes A-site interactions to perturb protein synthesis in bacteria.
Nat Commun, 14, 2023
|
|
9JUT
 
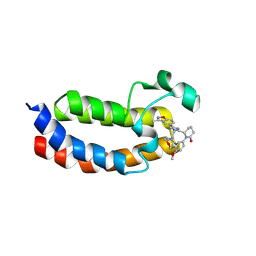 | | X-ray crystal structure of Y16524 in EP300 | | Descriptor: | (6~{S})-1-(3-chloranyl-4-methoxy-phenyl)-6-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{S})-2-morpholin-4-ylpropyl]imidazol-2-yl]piperidin-2-one, Histone acetyltransferase p300 | | Authors: | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | Deposit date: | 2024-10-08 | | Release date: | 2025-03-12 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
6LFX
 
 | |
7UUN
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with neomycin | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, NEOMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Osipiuk, J, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
9KNY
 
 | | Cryo-EM structure of pyruvate-treated human mitochondrial pyruvate carrier in the IMS-open conformation at pH 8.0 | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-11-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
6ARV
 
 | | Crystal structure of CARM1 with Compound 2 and SAH | | Descriptor: | (2R)-1-amino-3-{3-[4-(morpholin-4-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-b]pyridin-6-yl]phenoxy}propan-2-ol, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2017-08-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a CARM1 Inhibitor with Potent In Vitro and In Vivo Activity in Preclinical Models of Multiple Myeloma.
Sci Rep, 7, 2017
|
|
1KTT
 
 | | Thrombin inhibitor complex | | Descriptor: | 4-(5-BENZENESULFONYLAMINO-1-METHYL-1H-BENZOIMIDAZOL-2-YLMETHYL)-BENZAMIDINE, hirudin IIB, thrombin | | Authors: | Nar, H. | | Deposit date: | 2002-01-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of novel potent nonpeptide thrombin inhibitors.
J.Med.Chem., 45, 2002
|
|
4JYU
 
 | |
5FPY
 
 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 5-bromo-1-methyl-1H-indole-2-carboxylic acid (AT21457) in an alternate binding site. | | Descriptor: | 5-bromo-1-methyl-1H-indole-2-carboxylic acid, SERINE PROTEASE NS3 | | Authors: | Davies, T.G, Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FOM
 
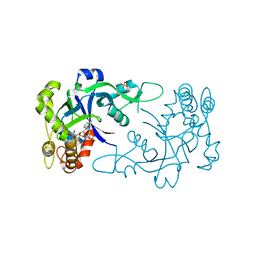 | | Crystal structure of the Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with the adduct AMP-AN6426 | | Descriptor: | 4-Chloro-3-aminomethyl-7-[ethoxy]-3H-benzo[C][1,2]oxaborol-1-ol modified adenosine, LEUCYL-TRNA SYNTHETASE, PHOSPHATE ION | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-24 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
7BO6
 
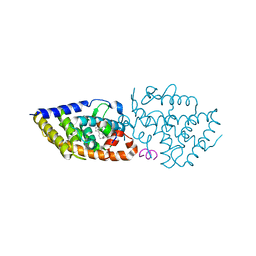 | | VDR complex with LCA derivative | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-01-24 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lithocholic acid-based design of noncalcemic vitamin D receptor agonists.
Bioorg.Chem., 111, 2021
|
|
3PYB
 
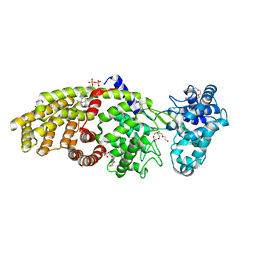 | | Crystal structure of ent-copalyl diphosphate synthase from Arabidopsis thaliana in complex with 13-aza-13,14-dihydrocopalyl diphosphate | | Descriptor: | 2-(methyl{2-[(1S,4aS,8aS)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]ethyl}amino)ethyl trihydrogen diphosphate, Ent-copalyl diphosphate synthase, chloroplastic, ... | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2010-12-12 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | Structure of ent-Copalyl Diphosphate Synthase from Arabidopsis thaliana, a Protonation-Dependent Diterpene Cyclase
To be Published
|
|
4Y4M
 
 | | Thiazole synthase Thi4 from Methanocaldococcus jannaschii | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Putative ribose 1,5-bisphosphate isomerase, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R)-2,3,5-tris(oxidanyl)-4-oxidanylidene-pentyl] hydrogen phosphate | | Authors: | Zhang, X, Ealick, S.E. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
6QQV
 
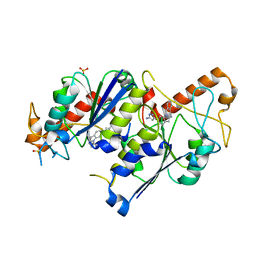 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 2-[[6-(5-azanyl-1~{H}-pyrazol-3-yl)indol-1-yl]methyl]benzenecarbonitrile, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
8DL2
 
 | | BoGH13ASus from Bacteroides ovatus bound to acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Brown, H.A, DeVeaux, A.L, Koropatkin, N.M. | | Deposit date: | 2022-07-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | BoGH13A Sus from Bacteroides ovatus represents a novel alpha-amylase used for Bacteroides starch breakdown in the human gut.
Cell.Mol.Life Sci., 80, 2023
|
|
6QR9
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-[[4-(morpholin-4-ylmethyl)phenyl]methyl]indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
9CT8
 
 | | Tricomplex of Compound 2, KRAS G12D, and CypA | | Descriptor: | (2R)-2-amino-N-(2-{[(1R)-1-cyclopentyl-2-{[(1M,8S,10S,14S,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]amino}-2-oxoethyl](methyl)amino}-2-oxoethyl)-N-methylpropanamide (non-preferred name), CHLORIDE ION, Isoform 2B of GTPase KRas, ... | | Authors: | Zhang, D, Bar Ziv, T, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-07-24 | | Release date: | 2025-07-23 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A neomorphic protein interface catalyzes covalent inhibition of RAS G12D aspartic acid in tumors.
Science, 389, 2025
|
|
3IJZ
 
 | | Lactobacillus casei Thymidylate Synthase ternary complex with dUMP and Pthalimidic derivative 15C | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(4-acetylphenyl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Pozzi, C, Cancian, L, Leone, R, Luciani, R, Ferrari, S, Mangani, S, Costi, M.P. | | Deposit date: | 2009-08-05 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
